Insights into dynamics of mobile genetic elements in hyperthermophilic environments from five new Thermococcus plasmids - PubMed (original) (raw)
Insights into dynamics of mobile genetic elements in hyperthermophilic environments from five new Thermococcus plasmids
Mart Krupovic et al. PLoS One. 2013.
Abstract
Mobilome of hyperthermophilic archaea dwelling in deep-sea hydrothermal vents is poorly characterized. To gain insight into genetic diversity and dynamics of mobile genetic elements in these environments we have sequenced five new plasmids from different Thermococcus strains that have been isolated from geographically remote hydrothermal vents. The plasmids were ascribed to two subfamilies, pTN2-like and pEXT9a-like. Gene content and phylogenetic analyses illuminated a robust connection between pTN2-like plasmids and Pyrococcus abyssi virus 1 (PAV1), with roughly half of the viral genome being composed of genes that have homologues in plasmids. Unexpectedly, pEXT9a-like plasmids were found to be closely related to the previously sequenced plasmid pMETVU01 from Methanocaldococcus vulcanius M7. Our data suggests that the latter observation is most compatible with an unprecedented horizontal transfer of a pEXT9a-like plasmid from Thermococcales to Methanococcales. Gene content analysis revealed that thermococcal plasmids encode Hfq-like proteins and toxin-antitoxin (TA) systems of two different families, VapBC and RelBE. Notably, although abundant in archaeal genomes, to our knowledge, TA and hfq-like genes have not been previously found in archaeal plasmids or viruses. Finally, the plasmids described here might prove to be useful in developing new genetic tools for hyperthermophiles.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
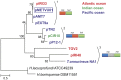
Figure 1. Phylogenetic tree of Thermococcales based on 16S rRNA gene sequences.
The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura-Nei model. The bootstrap consensus tree inferred from 100 replicates. A discrete Gamma distribution was used to model evolutionary rate differences among sites (4 categories (+G, parameter = 0.1068)). The rate variation model allowed for some sites to be evolutionarily invariable ([+_I_], 75.0448% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 45 nucleotide sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 1232 positions in the final dataset. Strains carrying sequenced plasmids are indicated by bold characters. The five plasmid-carrying Thermococcus strains isolated and characterized in this study are underlined.
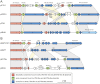
Figure 2. Comparative genomic analysis of the five novel thermococcal plasmids.
(A) pTN2-like and (B) pEXT9a-like plasmids Genes absolutely conserved in all eight plasmids are shown in red; the semi-conserved genes present in some members of both plasmid groups are in green; conserved genes restricted to either one of the two groups are depicted in blue. Abbreviations: SFI, superfamily I; wHTH, winged helix-turn-helix motif; ABC, ATP-binding cassette; RHH, ribbon-helix-helix motif; Prim-pol, primase-polymerase; TAS, toxin-antitoxin system. General characteristics of the plasmids depicted in this Figure can be found in Table 1.
Figure 3. Modular relationship among plasmid-encoded replication proteins.
Horizontal bars represent replication proteins with distinct domains indicated with different colours – the primase-polymerase domain is shown in blue, the C-terminal domain of pTN2 replicase is in red, and the winged helix-turn-helix domain is in green. The numbers above the bars represent the amino acid coordinates of the depicted domains in the replicases of respective plasmids (plasmid names are on the right). The numbers next to the plasmid names denote how many additional plasmids encode the specific type of a replication protein.
Figure 4. Phylogenetic analysis of the plasmid-encoded superfamily I helicases.
The plasmid names are coloured according to the geographical origin of the strains from which they were isolated: blue, East Pacific Ocean ridge; red, Mid-Atlantic Ocean ridge; green, Indian Ocean triple junction. Replicase–helicase (rep and hel) gene cassettes are shown on the right; domain organizations of the replication proteins encoded by plasmids within the three phylogenetic clades (1–3) are indicated. The plasmid of Methanocaldococcus vulcanius M7 is boxed. The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT model (+I, +G [4 categories]). Numbers at the branch-points represent bootstrap values (100 replicates). The scale bar represents the number of substitutions per site. The analysis involved 12 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 398 positions in the final dataset. The tree was rooted with helicases from haloarchaea, based on previous phylogenetic analysis . GenBank accession numbers: Halorubrum lacusprofundi ATCC 49239, YP_002564878; Halogeometricum borinquense DSM 11551, YP_004037766; Thermococcus onnurineus NA1, YP_002307767; Thermococcus gammatolerans EJ3 integrating element TGV2, YP_002959996; pMETVU01, YP_003248093; pTN2, YP_003603582; pP12-1, YP_003603439.
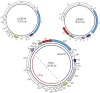
Figure 5. Relationship between thermococcal plasmids and virus PAV1.
Homologous genes are coloured similarly. PAV1 ORF528, which has homologues in haloarchaeal plasmids, is shaded grey.
Similar articles
- Two novel families of plasmids from hyperthermophilic archaea encoding new families of replication proteins.
Soler N, Marguet E, Cortez D, Desnoues N, Keller J, van Tilbeurgh H, Sezonov G, Forterre P. Soler N, et al. Nucleic Acids Res. 2010 Aug;38(15):5088-104. doi: 10.1093/nar/gkq236. Epub 2010 Apr 18. Nucleic Acids Res. 2010. PMID: 20403814 Free PMC article. - Molecular diversity of new Thermococcales isolates from a single area of hydrothermal deep-sea vents as revealed by randomly amplified polymorphic DNA fingerprinting and 16S rRNA gene sequence analysis.
Lepage E, Marguet E, Geslin C, Matte-Tailliez O, Zillig W, Forterre P, Tailliez P. Lepage E, et al. Appl Environ Microbiol. 2004 Mar;70(3):1277-86. doi: 10.1128/AEM.70.3.1277-1286.2004. Appl Environ Microbiol. 2004. PMID: 15006744 Free PMC article. - Diversity of CRISPR systems in the euryarchaeal Pyrococcales.
Norais C, Moisan A, Gaspin C, Clouet-d'Orval B. Norais C, et al. RNA Biol. 2013 May;10(5):659-70. doi: 10.4161/rna.23927. Epub 2013 Feb 19. RNA Biol. 2013. PMID: 23422322 Free PMC article. Review. - CRISPR elements in the Thermococcales: evidence for associated horizontal gene transfer in Pyrococcus furiosus.
Portillo MC, Gonzalez JM. Portillo MC, et al. J Appl Genet. 2009;50(4):421-30. doi: 10.1007/BF03195703. J Appl Genet. 2009. PMID: 19875895 - Plasmids, viruses and virus-like membrane vesicles from Thermococcales.
Soler N, Gaudin M, Marguet E, Forterre P. Soler N, et al. Biochem Soc Trans. 2011 Jan;39(1):36-44. doi: 10.1042/BST0390036. Biochem Soc Trans. 2011. PMID: 21265744 Review.
Cited by
- Thermococcus indicus sp. nov., a Fe(III)-reducing hyperthermophilic archaeon isolated from the Onnuri Vent Field of the Central Indian Ocean ridge.
Lim JK, Kim YJ, Yang JA, Namirimu T, Yang SH, Park MJ, Kwon YM, Lee HS, Kang SG, Lee JH, Kwon KK. Lim JK, et al. J Microbiol. 2020 Apr;58(4):260-267. doi: 10.1007/s12275-020-9424-9. Epub 2020 Apr 1. J Microbiol. 2020. PMID: 32239454 - Human endogenous retrovirus protein activates innate immunity and promotes experimental allergic encephalomyelitis in mice.
Perron H, Dougier-Reynaud HL, Lomparski C, Popa I, Firouzi R, Bertrand JB, Marusic S, Portoukalian J, Jouvin-Marche E, Villiers CL, Touraine JL, Marche PN. Perron H, et al. PLoS One. 2013 Dec 6;8(12):e80128. doi: 10.1371/journal.pone.0080128. eCollection 2013. PLoS One. 2013. PMID: 24324591 Free PMC article. - Unification of the globally distributed spindle-shaped viruses of the Archaea.
Krupovic M, Quemin ER, Bamford DH, Forterre P, Prangishvili D. Krupovic M, et al. J Virol. 2014 Feb;88(4):2354-8. doi: 10.1128/JVI.02941-13. Epub 2013 Dec 11. J Virol. 2014. PMID: 24335300 Free PMC article. - Extracellular membrane vesicles and nanotubes in Archaea.
Liu J, Soler N, Gorlas A, Cvirkaite-Krupovic V, Krupovic M, Forterre P. Liu J, et al. Microlife. 2021 Jun 10;2:uqab007. doi: 10.1093/femsml/uqab007. eCollection 2021. Microlife. 2021. PMID: 37223257 Free PMC article. Review. - Viruses Defined by the Position of the Virosphere within the Replicator Space.
Koonin EV, Dolja VV, Krupovic M, Kuhn JH. Koonin EV, et al. Microbiol Mol Biol Rev. 2021 Dec 15;85(4):e0019320. doi: 10.1128/MMBR.00193-20. Epub 2021 Sep 1. Microbiol Mol Biol Rev. 2021. PMID: 34468181 Free PMC article.
References
- Prieur D, Voytek MA, Jeanthon C, Reysenbach AL (2001) Deep-Sea Thermophilic Prokaryotes. In: Reysenbach AL, Voytek MA, Mancinelli R, editors. Thermophiles: Biodiversity, Ecology and Evolution. New York: Kluwer Academic/Plenum Publishers. pp. 11–22.
- Benbouzid-Rollet N, Lopez-Garcia P, Watrin L, Erauso G, Prieur D, et al. (1997) Isolation of new plasmids from hyperthermophilic Archaea of the order Thermococcales. Res Microbiol 148: 767–775. - PubMed
- Lepage E, Marguet E, Geslin C, Matte-Tailliez O, Zillig W, et al. (2004) Molecular diversity of new Thermococcales isolates from a single area of hydrothermal deep-sea vents as revealed by randomly amplified polymorphic DNA fingerprinting and 16S rRNA gene sequence analysis. Appl Environ Microbiol 70: 1277–1286. - PMC - PubMed
- Prieur D, Erauso G, Geslin C, Lucas S, Gaillard M, et al. (2004) Genetic elements of Thermococcales. Biochem Soc Trans 32: 184–187. - PubMed
- Soler N, Gaudin M, Marguet E, Forterre P (2011) Plasmids, viruses and virus-like membrane vesicles from Thermococcales. Biochem Soc Trans 39: 36–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This work was supported by the CNRS, the MESR, the IFREMER and grants from the EU projects PYRED QLK36CT-2001-01676 to GE and Marine Genomic Europe MGE GOCE-CT-2004-5054003, ANR Genoarchaea M2TFP and the Souchothèque de Bretagne (Brittany Microbe Culture Collection). MG has received a PhD Fellowship from the Région Bretagne. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials