Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment - PubMed (original) (raw)
doi: 10.1371/journal.pone.0053028. Epub 2013 Jan 9.
Wenke Feng, Irina Kirpich, Yuhua Wang, Xiang Qin, Yanlong Liu, Leila Gobejishvili, Swati Joshi-Barve, Tulin Ayvaz, Joseph Petrosino, Maiying Kong, David Barker, Craig McClain, Shirish Barve
Affiliations
- PMID: 23326376
- PMCID: PMC3541399
- DOI: 10.1371/journal.pone.0053028
Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment
Lara Bull-Otterson et al. PLoS One. 2013.
Abstract
Enteric dysbiosis plays an essential role in the pathogenesis of alcoholic liver disease (ALD). Detailed characterization of the alterations in the gut microbiome is needed for understanding their pathogenic role in ALD and developing effective therapeutic approaches using probiotic supplementation. Mice were fed liquid Lieber-DeCarli diet without or with alcohol (5% v/v) for 6 weeks. A subset of mice were administered the probiotic Lactobacillus rhamnosus GG (LGG) from 6 to 8 weeks. Indicators of intestinal permeability, hepatic steatosis, inflammation and injury were evaluated. Metagenomic analysis of the gut microbiome was performed by analyzing the fecal DNA by amplification of the V3-V5 regions of the 16S rRNA gene and large-scale parallel pyrosequencing on the 454 FLX Titanium platform. Chronic ethanol feeding caused a decline in the abundance of both Bacteriodetes and Firmicutes phyla, with a proportional increase in the gram negative Proteobacteria and gram positive Actinobacteria phyla; the bacterial genera that showed the biggest expansion were the gram negative alkaline tolerant Alcaligenes and gram positive Corynebacterium. Commensurate with the qualitative and quantitative alterations in the microbiome, ethanol caused an increase in plasma endotoxin, fecal pH, hepatic inflammation and injury. Notably, the ethanol-induced pathogenic changes in the microbiome and the liver were prevented by LGG supplementation. Overall, significant alterations in the gut microbiome over time occur in response to chronic alcohol exposure and correspond to increases in intestinal barrier dysfunction and development of ALD. Moreover, the altered bacterial communities of the gut may serve as significant therapeutic target for the prevention/treatment of chronic alcohol intake induced intestinal barrier dysfunction and liver disease.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
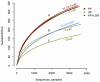
Figure 1. Rarefaction curves indicating the observed number of operational taxonomic units (OTUs) within a sample.
Rarefaction curves of OTUs clustered and saturated at different level across exposure groups and indicate the intra-sample diversity. While the baseline PF and AF OTUs cluster together with the PF T2 (end of week 6) and PF T3 (end of week 8) groups (A), the AF T2 and LGG groups (B) saturate earlier than the baseline group show they are less diverse than the baseline group. The AF T3 exposure group (C) saturates the earliest, indicating it is the least diverse sample.
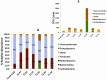
Figure 2. The relative abundance of bacteria phylum by exposure group over time.
Time is indicted by time one -T1 (baseline), time two = T2 (end of week 6), time three - T3 (end of week 8). The phylum abundance is indicated by the color bars. The exact percentages of Bacteroidetes’ and Firmicutes’ relative abundance are shown in 2A and given by the numbers on the bars. The relative abundance of less common bacterial taxa found after removal of Bacteroidetes and Firmicutes are shown in 2B. The microbiomes of mice exposed to alcohol at T2 and T3 are characterized by greater abundance of Proteobacteria and Actinobacteria.
Figure 3. Weighted PCoA of bacterial genus by time and exposure group using UniFrac distance matrix.
This dimension reduction analysis demonstrates the clustering of samples based on their location and similar microbial profiles. The first component explains 69% and the second component explains 13% of the variance in the samples. AF T3 group is the most divergent of the exposure groups while AF T2 and AF+LGG T3 are separated from the other baseline samples as well. AF T1, PF T1 are most similar as are PF T2 and T3.
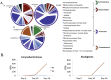
Figure 4. The relative distribution of the bacterial phyla and genera in response to ethanol feeding and LGG supplementation.
(A) The microbiome of the PF (right), AF+LGG (left) and AF (middle lower) groups at T3 are shown in the pie charts and color coordinated by genus and phylum. The different shades of color represent the different genera and the common color spectrum (reds, purples, green and orange) represents the phyla. The outer ring around the pie charts also depicts the different phyla. The microbiomes of mice exposed to AF are characterized by greater abundance of Alcaligenes and Corynebacterium and loss of Tannerella. The AF+LGG group shows a much greater abundance of Lactobacillus and nonspecific Ruminococcaceae Incertae Sedis compared to the other exposure groups. (B) The growth rate over time of Alcaligenes and Corynebacterium in the PF and AF groups shows the expansion of the genera after exposure to alcohol.
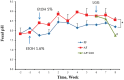
Figure 5. Fecal pH changes in response to alcohol feeding and LGG supplementation.
Animals were fed Lieber-DeCarli liquid diet with alcohol (AF) or without (PF). LGG supplementation (AF+LGG) was started at the end of week 6 and continued till the end of the experiment (8 weeks). Fecal samples were collected weekly. Data was obtained from n = 6–8 for the PF and AF groups and n = 4 for the AF+LGG group. Fecal pH significantly increased in response to alcohol feeding over the study period of 8 weeks (*p<0.0001, AF versus PF). Comparison of fecal samples obtained from LGG supplemented animals (AF+LGG) with alcohol fed animals (AF) obtained at the end of the study period (samples at the end of week 8) showed a significant decrease in fecal pH (# p = 0.001, AF versus AF+LGG at week 8).
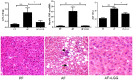
Figure 6. LGG probiotic supplementation ameliorates alcohol-induced blood endotoxemia, hepatic inflammation, steatosis and injury.
Data was obtained from n = 6–8 for the PF and AF groups and n = 4 for the AF+LGG group. (A) Plasma endotoxin levels assessed by LPS (lipopolysaccharides) measurement. Two weeks of LGG supplementation attenuated alcohol-mediated increase in plasma LPS levels. (B) Hepatic marker of inflammation, TNF-α m-RNA, was significantly up-regulated in response to alcohol feeding. LGG supplementation was able to bring it down to the level of control pair-fed animals. (C) Liver injury was evaluated by plasma ALT activity. Alcohol feeding resulted in significant elevation of plasma ALT levels compared to control pair-fed animals, which was decreased by LGG treatment. (D) Hematoxylineosin staining demonstrated severe micro- and macrovesicular fat accumulation in the livers of alcohol fed mice compared to control pair-fed animals. Two weeks of LGG treatment markedly attenuated alcohol-induced hepatic steatosis. Arrow indicates the fat droplets and arrow head denotes neutrophil infiltration foci (x200 final magnification). *p<0.05; **p<0.01; ***p<0.001.
Similar articles
- Lactobacillus rhamnosus GG reduces hepatic TNFα production and inflammation in chronic alcohol-induced liver injury.
Wang Y, Liu Y, Kirpich I, Ma Z, Wang C, Zhang M, Suttles J, McClain C, Feng W. Wang Y, et al. J Nutr Biochem. 2013 Sep;24(9):1609-15. doi: 10.1016/j.jnutbio.2013.02.001. Epub 2013 Apr 22. J Nutr Biochem. 2013. PMID: 23618528 Free PMC article. - Lactobacillus rhamnosus GG combined with inosine ameliorates alcohol-induced liver injury through regulation of intestinal barrier and Treg/Th1 cells.
Zhu Y, Wang X, Zhu L, Tu Y, Chen W, Gong L, Pan T, Lin H, Lin J, Sun H, Ge Y, Wei L, Guo Y, Lu C, Chen Y, Xu L. Zhu Y, et al. Toxicol Appl Pharmacol. 2022 Mar 15;439:115923. doi: 10.1016/j.taap.2022.115923. Epub 2022 Feb 15. Toxicol Appl Pharmacol. 2022. PMID: 35176292 - Supplementation of saturated long-chain fatty acids maintains intestinal eubiosis and reduces ethanol-induced liver injury in mice.
Chen P, Torralba M, Tan J, Embree M, Zengler K, Stärkel P, van Pijkeren JP, DePew J, Loomba R, Ho SB, Bajaj JS, Mutlu EA, Keshavarzian A, Tsukamoto H, Nelson KE, Fouts DE, Schnabl B. Chen P, et al. Gastroenterology. 2015 Jan;148(1):203-214.e16. doi: 10.1053/j.gastro.2014.09.014. Epub 2014 Sep 16. Gastroenterology. 2015. PMID: 25239591 Free PMC article. - Alcohol or Gut Microbiota: Who Is the Guilty?
Meroni M, Longo M, Dongiovanni P. Meroni M, et al. Int J Mol Sci. 2019 Sep 14;20(18):4568. doi: 10.3390/ijms20184568. Int J Mol Sci. 2019. PMID: 31540133 Free PMC article. Review. - Alcoholic liver disease: the gut microbiome and liver cross talk.
Hartmann P, Seebauer CT, Schnabl B. Hartmann P, et al. Alcohol Clin Exp Res. 2015 May;39(5):763-75. doi: 10.1111/acer.12704. Alcohol Clin Exp Res. 2015. PMID: 25872593 Free PMC article. Review.
Cited by
- The role of the gut-brain axis in alcohol use disorders.
Gorky J, Schwaber J. Gorky J, et al. Prog Neuropsychopharmacol Biol Psychiatry. 2016 Feb 4;65:234-41. doi: 10.1016/j.pnpbp.2015.06.013. Epub 2015 Jul 16. Prog Neuropsychopharmacol Biol Psychiatry. 2016. PMID: 26188287 Free PMC article. Review. - Oligofructose protects against arsenic-induced liver injury in a model of environment/obesity interaction.
Massey VL, Stocke KS, Schmidt RH, Tan M, Ajami N, Neal RE, Petrosino JF, Barve S, Arteel GE. Massey VL, et al. Toxicol Appl Pharmacol. 2015 May 1;284(3):304-14. doi: 10.1016/j.taap.2015.02.022. Epub 2015 Mar 8. Toxicol Appl Pharmacol. 2015. PMID: 25759243 Free PMC article. - Explaining diversity in metagenomic datasets by phylogenetic-based feature weighting.
Albanese D, De Filippo C, Cavalieri D, Donati C. Albanese D, et al. PLoS Comput Biol. 2015 Mar 27;11(3):e1004186. doi: 10.1371/journal.pcbi.1004186. eCollection 2015 Mar. PLoS Comput Biol. 2015. PMID: 25815895 Free PMC article. - Alcohol-Related Elevation of Liver Transaminase Is Associated With Gut Microbiota in Male.
Jiao M, Yan S, Shi Q, Liu Y, Li Y, Lv J, Ding S, Li A. Jiao M, et al. Front Med (Lausanne). 2022 Feb 22;9:823898. doi: 10.3389/fmed.2022.823898. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35280887 Free PMC article. - Probiotics attenuate alcohol-induced muscle mechanical hyperalgesia: Preliminary observations.
Green PG, Alvarez P, Levine JD. Green PG, et al. Mol Pain. 2022 Jan-Dec;18:17448069221075345. doi: 10.1177/17448069221075345. Mol Pain. 2022. PMID: 35189754 Free PMC article.
References
- Adachi Y, Moore LE, Bradford BU, Gao W, Thurman RG (1995) Antibiotics prevent liver injury in rats following long-term exposure to ethanol. Gastroenterology 108: 218–224. - PubMed
- Bode C, Kugler V, Bode JC (1987) Endotoxemia in patients with alcoholic and non-alcoholic cirrhosis and in subjects with no evidence of chronic liver disease following acute alcohol excess. Journal of hepatology 4: 8–14. - PubMed
- Fukui H, Brauner B, Bode JC, Bode C (1991) Plasma endotoxin concentrations in patients with alcoholic and non-alcoholic liver disease: reevaluation with an improved chromogenic assay. Journal of hepatology 12: 162–169. - PubMed
- Keshavarzian A, Choudhary S, Holmes EW, Yong S, Banan A, et al. (2001) Preventing gut leakiness by oats supplementation ameliorates alcohol-induced liver damage in rats. The Journal of pharmacology and experimental therapeutics 299: 442–448. - PubMed
- Nanji AA, Khettry U, Sadrzadeh SM (1994) Lactobacillus feeding reduces endotoxemia and severity of experimental alcoholic liver (disease). Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine 205: 243–247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 AA021901/AA/NIAAA NIH HHS/United States
- R01 AA018869/AA/NIAAA NIH HHS/United States
- RC2AA019385/AA/NIAAA NIH HHS/United States
- P30 AA019360/AA/NIAAA NIH HHS/United States
- R01 AA015970/AA/NIAAA NIH HHS/United States
- R01 AAO14371/PHS HHS/United States
- R01 AA014371/AA/NIAAA NIH HHS/United States
- R01 AA0015970/AA/NIAAA NIH HHS/United States
- R37 AA010762/AA/NIAAA NIH HHS/United States
- P01 AA017103/AA/NIAAA NIH HHS/United States
- R21 AA020848/AA/NIAAA NIH HHS/United States
- R01 AA018016/AA/NIAAA NIH HHS/United States
- R21 AA020849/AA/NIAAA NIH HHS/United States
- R01 DK071765/DK/NIDDK NIH HHS/United States
- RC2 AA019385/AA/NIAAA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources