RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection - PubMed (original) (raw)
RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection
J Ross Chapman et al. Mol Cell. 2013.
Erratum in
- RIF1 Is Essential for 53BP1-Dependent Nonhomologous End Joining and Suppression of DNA Double-Strand Break Resection.
Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ. Chapman JR, et al. Mol Cell. 2021 Jul 1;81(13):2868. doi: 10.1016/j.molcel.2021.06.015. Mol Cell. 2021. PMID: 34214445 Free PMC article. No abstract available.
Abstract
The appropriate execution of DNA double-strand break (DSB) repair is critical for genome stability and tumor avoidance. 53BP1 and BRCA1 directly influence DSB repair pathway choice by regulating 5' end resection, but how this is achieved remains uncertain. Here we report that Rif1(-/-) mice are severely compromised for 53BP1-dependent class switch recombination (CSR) and fusion of dysfunctional telomeres. The inappropriate accumulation of RIF1 at DSBs in S phase is antagonized by BRCA1, and deletion of Rif1 suppresses toxic nonhomologous end joining (NHEJ) induced by PARP inhibition in Brca1-deficient cells. Mechanistically, RIF1 is recruited to DSBs via the N-terminal phospho-SQ/TQ domain of 53BP1, and DSBs generated by ionizing radiation or during CSR are hyperresected in the absence of RIF1. Thus, RIF1 and 53BP1 cooperate to block DSB resection to promote NHEJ in G1, which is antagonized by BRCA1 in S phase to ensure a switch of DSB repair mode to homologous recombination.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
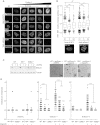
Defective CSR in RIF1_-_Deficient Mice (A) Quantification of immunoglobulins in the serum of Rif1+/+ and _Rif1_−/− mice. (B–D) Flow cytometry staining (B) and quantifications of surface expression (C) and secreted (D) IgG1 and IgG2b in Rif1 +/+ and _Rif1_−/− B cells stimulated with LPS or anti-mouse CD40 in the presence or absence of IL-4. (E) B cells were labeled with CTV and cultured for 3 days with anti-mouse CD40 and IL-4 (black line). Grey profile, unstimulated control. Numbers of cell divisions are depicted on the top of the graphs. (F) Rif1+/+ and _Rif1_−/− mice were immunized with NP-KLH, and NP-specific IgM (left) and IgG (middle) were measured in mice serum at different time points after immunization. Anti-NP-specific antibodies are shown for Rif1+/+ and _Rif1_−/− mice at 21 days after immunization (right). Error bars are ± SEM. See also Figures S1–S3 and Table S1.
Figure 2
A Requirement for RIF1 in the NHEJ of Dysfunctional Telomeres (A) Western blot showing comparable expression of indicated Trf2 transgenes between MEF lines of the indicated genotype. (B) Metaphases were analyzed for telomere end-to-end fusions in cells of indicated genotype, 96 hr following retroviral transduction of the indicated TRF2 expression constructs. Right panels show enlargement of corresponding left panel. Telomeric fluorescence in situ hybridization (FISH), red; 4,6-diamidino-2-phenylindole (DAPI), gray. (C) Quantification of chromosome fusions. n = 3,000, 2,000, and 3,000 chromosomes scored per genotype, over two independent experiments. ∗∗∗p < 0 .0001 (one-way ANOVA). Error bars are ± SEM.
Figure 3
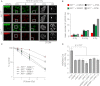
RIF1 Deficiency Suppresses Proliferation and Genome Instability Defects in BRCA1_-_Depleted Cells (A) HeLa cells were treated with the indicated siRNA for 48 hr. Cells were then pulse-labeled with EdU (5 min, 40 μM) immediately before irradiation (5 Gy). One hour following IR, cells were fixed and permeabilized before fluorescent labeling of EdU and counterstaining with indicated antisera. Representative projection images of whole-nuclei 0.5 μm confocal z series are presented. (B) Enlarged S phase images of RIF1 IRIF from (A) and automated quantification of the intensity of RIF1 IRIF in HeLa cells subjected to control or BRCA1-targeting siRNA. n > 140 cells per condition; ∗∗∗p < 0.0001, ns1 p = 0.514, ns2 p = 0.1172, ns3 p = 0.1390, Mann-Whitney test. (C) Western blot shows comparable BRCA1-depletion efficiency between MEF lines selected for expression of indicated shRNA constructs. C, control shRNA vector; 1, Brca1 shRNA vector 1; 2, Brca1 shRNA vector 2. (D) Radial chromosomes were scored in metaphases prepared from WT, Rif1 −/−, and 53Bp1 −/− cell lines expressing the indicated shRNA following 16 hr control or olaparib (1 μM) treatments. n = 50 metaphase per condition. Error bars are ± SEM. See also Figure S4.
Figure 4
A Role for RIF1 in the Cellular Responses to DSBs (A) WT and Rif1 −/− MEFs were untreated (CNTL) or harvested at indicated times following γ-irradiation before immunostaining with the indicated antisera. Zoom panels correspond to indicated cells in left panels. (B) γH2AX IRIF were scored in cells of indicated genotype 24 hr following mock treatment or IR at the indicated doses. Where indicated, cells were either mock treated (DMSO) or incubated with ATMi (KU55933, 10 μM) from 30 min before irradiation until harvesting. Mean number of γH2AX IRIF per cell is plotted ± SEM. n > 200 cells/condition over two independent experiments. (C) The survival of MEFs of indicated genotype following control or γ-irradiation treatments was assessed by colony survival assay. n = 3 ± SEM. (D) Measurement of NHEJ proficiency in HEK239 EJ5 cells subjected to the indicated siRNAs. n = 3 ± SEM unpaired two-tailed t test. See also Figure S5.
Figure 5
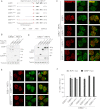
RIF1 Recruitment to DNA Damage Sites Requires Phosphorylation of the of 53BP1 S/TQ Domain (A) Schematic of 53BP1 mutants generated to examine the requirements for RIF1 IRIF. Numbers indicate N- and C-terminal 53BP1 residues for each truncation mutant. Lines indicate relative positions of S/T residues in each S/TQ motif and whether they were either alanine-substituted (red) or left intact (blue) in each respective mutant. (B) Western blot demonstrating comparable expression of hemagglutinin (HA)-tagged 53BP1 mutants analyzed in (C) upon lentivirus-mediated reconstitution in 53Bp1 −/− cells. (C) The ability of the indicated 53BP1 mutant to support RIF1 IRIF was examined by indirect immunofluorescence in irradiated cells 1 hr following IR. (D) Western blot of the indicated 53BP1 mutants reconstituted in 53Bp1 −/− cells and analyzed in (E) to examine the function of the 53BP1 N terminus and its S/TQ ATM consensus phosphorylation in promoting RIF1 IRIF. (E) Similar to (C) but with the indicated 53BP1 mutants reconstituted in 53Bp1 −/− cells. Arrowheads indicate colocalizing foci. (F) Quantification of RIF1 and 53BP1 IRIF with the indicated 53BP1 mutants reconstituted in 53Bp1 −/− cells. n = 2, >140 cells scored per condition ± SEM.
Figure 6
ATM-Dependent Phosphorylation of 53BP1 Promotes RIF1 Interaction (A) Irradiated WT MEFs pretreated with DMSO or ATMi were fixed and immunostained for γH2AX and either RIF1 or 53BP1. (B) Quantification of 53BP1 and RIF1 IRIF in cells prepared as in (A). n > 4 ± SD, >126 cells scored per condition. (C) Immunoblots of whole-cell lysates prepared from 53Bp1 −/− MEFs complemented with the indicated constructs as in Figure 5 1 hr following mock or 10 Gy IR. (D) Flag-HA-53BP1 proteins were purified with Flag-M2 beads (Sigma) from lysates in (C) under stringent conditions. Bead complexes were subsequently incubated in nuclear extracts (HNE) before 53BP1-associated proteins were recovered by peptide elution. B/O = beads-only control. (E) Immunoblots of whole-cell lysates of 53Bp1 −/− MEFs complemented with WT 53BP1, 1 hr following mock or 15 Gy IR treatments in the presence/absence of ATMi. (F) As in (D), but with FLAG-HA-53BP1 purified from lysates indicated in (E). See also Figure S6.
Figure 7
RIF1 Represses DSB Resection Following IR and at the IgH Locus in Stimulated B Cells (A and B) Enhanced ATR signaling in RIF1- and 53BP1_-_deficent cells. Lysates prepared from WT, Rif1 −/−, and 53Bp1 −/− MEFs, harvested at indicated time points following IR, were immunoblotted with indicated antisera. (C) 53Bp1 −/− MEF lines reconstituted with indicated WT and mutant 53BP1 proteins (see also Figures 5A–5D) were examined for IR-induced checkpoint phosphorylation events as in (A) and (B). (D) Lysates prepared from WT, Rif1 −/−, 53Bp1 −/−, and Rif1 −/− 53Bp1 −/− MEFs harvested 3 hr following IR were immunoblotted with indicated antisera. (E) Aberrant processing of the IgH locus in Rif1 −/− cells. Schematic of IgH Sμ region shows relative positions of qPCR amplicons used in ChIP experiments. A control non-IgH locus (Rpp30) was also examined. Resting B cells or those stimulated with LPS and IL-4 (72 hr) were subjected to ChIP experiments with IgG (control), histone H2A.X, and RPA32 monoclonal antisera. Following background subtraction of IgG signals, values were normalized to the DNA input signals, followed by the maximum value in each data set. Mean signals, two independent experiments ± SEM. See also Figure S7.
Comment in
- RIF1 in DNA break repair pathway choice.
Daley JM, Sung P. Daley JM, et al. Mol Cell. 2013 Mar 7;49(5):840-1. doi: 10.1016/j.molcel.2013.02.019. Mol Cell. 2013. PMID: 23473603 Free PMC article.
Similar articles
- 53BP1 regulates DSB repair using Rif1 to control 5' end resection.
Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. Zimmermann M, et al. Science. 2013 Feb 8;339(6120):700-4. doi: 10.1126/science.1231573. Epub 2013 Jan 10. Science. 2013. PMID: 23306437 Free PMC article. - Impaired 53BP1/RIF1 DSB mediated end-protection stimulates CtIP-dependent end resection and switches the repair to PARP1-dependent end joining in G1.
Bakr A, Köcher S, Volquardsen J, Petersen C, Borgmann K, Dikomey E, Rothkamm K, Mansour WY. Bakr A, et al. Oncotarget. 2016 Sep 6;7(36):57679-57693. doi: 10.18632/oncotarget.11023. Oncotarget. 2016. PMID: 27494840 Free PMC article. - 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions.
Callen E, Di Virgilio M, Kruhlak MJ, Nieto-Soler M, Wong N, Chen HT, Faryabi RB, Polato F, Santos M, Starnes LM, Wesemann DR, Lee JE, Tubbs A, Sleckman BP, Daniel JA, Ge K, Alt FW, Fernandez-Capetillo O, Nussenzweig MC, Nussenzweig A. Callen E, et al. Cell. 2013 Jun 6;153(6):1266-80. doi: 10.1016/j.cell.2013.05.023. Epub 2013 May 30. Cell. 2013. PMID: 23727112 Free PMC article. - Roles for 53BP1 in the repair of radiation-induced DNA double strand breaks.
Shibata A, Jeggo PA. Shibata A, et al. DNA Repair (Amst). 2020 Sep;93:102915. doi: 10.1016/j.dnarep.2020.102915. DNA Repair (Amst). 2020. PMID: 33087281 Review. - Regulation of repair pathway choice at two-ended DNA double-strand breaks.
Shibata A. Shibata A. Mutat Res. 2017 Oct;803-805:51-55. doi: 10.1016/j.mrfmmm.2017.07.011. Epub 2017 Jul 29. Mutat Res. 2017. PMID: 28781144 Review.
Cited by
- Interaction of BARD1 and HP1 Is Required for BRCA1 Retention at Sites of DNA Damage.
Wu W, Nishikawa H, Fukuda T, Vittal V, Asano M, Miyoshi Y, Klevit RE, Ohta T. Wu W, et al. Cancer Res. 2015 Apr 1;75(7):1311-21. doi: 10.1158/0008-5472.CAN-14-2796. Epub 2015 Jan 29. Cancer Res. 2015. PMID: 25634209 Free PMC article. - Replication fork stability confers chemoresistance in BRCA-deficient cells.
Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, Lee JE, Wong N, Lafarga V, Calvo JA, Panzarino NJ, John S, Day A, Crespo AV, Shen B, Starnes LM, de Ruiter JR, Daniel JA, Konstantinopoulos PA, Cortez D, Cantor SB, Fernandez-Capetillo O, Ge K, Jonkers J, Rottenberg S, Sharan SK, Nussenzweig A. Ray Chaudhuri A, et al. Nature. 2016 Jul 21;535(7612):382-7. doi: 10.1038/nature18325. Nature. 2016. PMID: 27443740 Free PMC article. - 53BP1 Integrates DNA Repair and p53-Dependent Cell Fate Decisions via Distinct Mechanisms.
Cuella-Martin R, Oliveira C, Lockstone HE, Snellenberg S, Grolmusova N, Chapman JR. Cuella-Martin R, et al. Mol Cell. 2016 Oct 6;64(1):51-64. doi: 10.1016/j.molcel.2016.08.002. Epub 2016 Aug 18. Mol Cell. 2016. PMID: 27546791 Free PMC article. - 53BP1 fosters fidelity of homology-directed DNA repair.
Ochs F, Somyajit K, Altmeyer M, Rask MB, Lukas J, Lukas C. Ochs F, et al. Nat Struct Mol Biol. 2016 Aug;23(8):714-21. doi: 10.1038/nsmb.3251. Epub 2016 Jun 27. Nat Struct Mol Biol. 2016. PMID: 27348077 - Checkpoint phosphorylation sites on budding yeast Rif1 protect nascent DNA from degradation by Sgs1-Dna2.
Gali VK, Monerawela C, Laksir Y, Hiraga SI, Donaldson AD. Gali VK, et al. PLoS Genet. 2023 Nov 13;19(11):e1011044. doi: 10.1371/journal.pgen.1011044. eCollection 2023 Nov. PLoS Genet. 2023. PMID: 37956214 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous