Pharmacological modulation of circadian rhythms by synthetic activators of the deacetylase SIRT1 - PubMed (original) (raw)
. 2013 Feb 26;110(9):3333-8.
doi: 10.1073/pnas.1214266110. Epub 2013 Jan 22.
Yasukazu Nakahata, Mohamed Boudjelal, Emma Watts, Danuta E Mossakowska, Kenneth A Edwards, Marlene Cervantes, Giuseppe Astarita, Christine Loh, James L Ellis, George P Vlasuk, Paolo Sassone-Corsi
Affiliations
- PMID: 23341587
- PMCID: PMC3587185
- DOI: 10.1073/pnas.1214266110
Pharmacological modulation of circadian rhythms by synthetic activators of the deacetylase SIRT1
Marina M Bellet et al. Proc Natl Acad Sci U S A. 2013.
Abstract
Circadian rhythms govern a wide variety of physiological and metabolic functions in many organisms, from prokaryotes to humans. We previously reported that silent information regulator 1 (SIRT1), a NAD(+)-dependent deacetylase, contributes to circadian control. In addition, SIRT1 activity is regulated in a cyclic manner in virtue of the circadian oscillation of the coenzyme NAD(+). Here we used specific SIRT1 activator compounds both in vitro and in vivo. We tested a variety of compounds to show that the activation of SIRT1 alters CLOCK:BMAL1-driven transcription in different systems. Activation of SIRT1 induces repression of circadian gene expression and decreases H3 K9/K14 acetylation at corresponding promoters in a time-specific manner. Specific activation of SIRT1 was demonstrated in vivo using liver-specific SIRT1-deficient mice, where the effect of SIRT1 activator compounds was shown to be dependent on SIRT1. Our findings demonstrate that SIRT1 can fine-tune circadian rhythm and pave the way to the development of pharmacological strategies to address a broad range of therapeutic indications.
Conflict of interest statement
Conflict of interest statement: M.B., E.W., D.E.M., K.A.E., C.L., J.L.E., and G.P.V. are employees of GlaxoSmithKline. P.S.-C. is a member of the Sirtris Scientific Advisory Board.
Figures
Fig. 1.
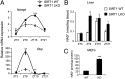
Absence of SIRT1 alters NAD+ cellular levels. (A) Livers from SIRT1Δex4 mice (LKO) and isogenic WT were collected at four different times of the circadian cycle. Nampt and Dbp mRNA expression profiles were analyzed by quantitative PCR. The values are relative to those of 18S mRNA levels at each ZT. All values are the mean ± SEM (n = 3); **P < 0.01, ***P < 0.001. (B) NAD+ was extracted from WT and SIRT1Δex4 livers (LKO) at the indicated time points and analyzed by LC/MSn. Results were normalized for mg of liver tissue. All values are the mean ± SD (n = 3); *P < 0.05. (C) Cellular NAD+ was extracted from WT and _Sirt1_−/− MEFs and analyzed by LC/MSn. Results were normalized for micrograms of protein. All values are the mean ± SD (n = 3); ***P < 0.001.
Fig. 2.
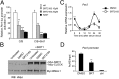
Modulation of SIRT1 alters CLOCK:BMAL1-mediated transcription. (A) JEG3 cells were transfected with 50 ng of m_Per1_-luciferase promoter together with CLOCK (C; 100 ng) and BMAL1 (B; 50 ng), with or without SIRT1 (100 ng). Six hours after transfection, cells were treated with SRT2183 10 and 20 μM or with NAD+ 1 mM. DMSO treatment was added in control cells. The effect of the treatment on CLOCK:BMAL1-induced transcription was evaluated 24 h after transfection. After normalization for transfection efficiency using β-galactosidase activity, reporter gene activities were expressed as percentage of relative activation. Value of transcriptional activation in controls (ctrl) was set to 100%. All of the values are the mean ± SD (n = 3); **P < 0.01, ***P < 0.001. (B) Immunoblot analysis of protein extracts obtained from HEK 293 cells transfected with CLOCK:BMAL1 expression vectors alone or with SIRT1. Six hours after transfection, cells were left untreated or treated with DMSO or SRT2183 for 8 h. Nuclear extracts were prepared and loaded on a 6% acrylamide gel. CLOCK and BMAL1 amount was evaluated using Myc antibody. (C) WT MEFs were pretreated with SRT2183 10 μM or DMSO for 16 h before serum shock. Per2 mRNA expression profile was analyzed by quantitative PCR. The values are relative to those of 18S mRNA levels at each circadian time. All of the values are the mean ± SEM (n = 3); *P < 0.05, **P < 0.01, ***P < 0.001. (D) Histone H3 (K9/K14) acetylation at Per2 promoter. WT MEFs were treated with SRT2183 10 μM at time 15 (CT15) after serum shock for 1 h. Cross-linked cell extracts were isolated and subjected to ChIP analysis with antiacetyl histone H3 (K9/K14) and control IgG (ctrl), and analyzed by quantitative PCR. All of the values are the mean ± SD (n = 3); **P < 0.01.
Fig. 3.
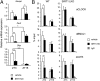
Modification of circadian expression in WT mice treated with SRT1720. (A) Change in circadian expression of Nampt, Per2, and Dbp clock genes in livers from mice treated with SRT1720 for 3 wk, compared with vehicle-treated mice. The values are relative to those of 18S mRNA levels at each ZT. Three animals for each condition were used. All of the values are the mean ± SD (n = 3); *P < 0.05, **P < 0.01, ***P < 0.001. (B) ChIP from livers of WT and SIRT1Δex4 mice (LKO) treated with SRT1720 or with vehicle for 3 wk. Livers were collected at two ZT (ZT3 and ZT15) and subjected to dual cross-link and DNA was immunoprecipitated with anti-CLOCK, anti-BMAL1, anti-H3(Lys9/Lys14), and rabbit IgG. Primers for Dbp promoter UP region were used for quantitative PCR. All of the values are the mean ± SD (n = 3); *P < 0.05, **P < 0.01.
Fig. 4.
SIRT1 activators decrease the amplitude of the bioluminescence oscillations. U2OS cells stably transfected with _Bmal1_-luciferase promoter (U2OS-h-Bmal) and NIH 3T3 stably transfected with _Per2_-luciferase promoter (NIH-h-Per2) were synchronized by Dex treatment and the bioluminescence was red every 90 min for 5 consecutive days in DMSO-treated cells (black line) compared with cells treated with SRTCE1022, CD1023, CL1015 (red line) at the concentration of 1 μM and 10 μM.
Fig. 5.
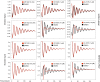
SIRT1 activators repress circadian gene expression in a SIRT1-specific manner. (A) Per2, Dbp, and Cry1 mRNA expression profiles in WT MEFs pretreated with DMSO, SRT CD1023, CL1015, and CE1022 (10 μM), before serum shock synchronization, were analyzed by quantitative PCR. The values are relative to those of Gapdh mRNA levels at each circadian time. Time 0 value in DMSO-treated cells was set to 1. All of the values are the mean ± SEM (n = 3); *P < 0.05, **P < 0.01. (B) Cross-linked cell extract were isolated at the indicated time points after serum shock from WT MEFs pretreated with SRTCD1023, CL1015, and CE1022 (10 μM). The samples were subjected to ChIP assay with anti-CLOCK and anti-IgG (ctr), and analyzed by quantitative PCR with primers for Per2 and Dbp promoter. All of the values are the mean ± SD (n = 3); *P < 0.05, **P < 0.01, ***P < 0.001. (C) Same experimental conditions as in B, but using SIRT1−/− MEFs.
Similar articles
- The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control.
Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. Nakahata Y, et al. Cell. 2008 Jul 25;134(2):329-40. doi: 10.1016/j.cell.2008.07.002. Cell. 2008. PMID: 18662547 Free PMC article. - CLOCK and BMAL1 Regulate Muscle Insulin Sensitivity via SIRT1 in Male Mice.
Liu J, Zhou B, Yan M, Huang R, Wang Y, He Z, Yang Y, Dai C, Wang Y, Zhang F, Zhai Q. Liu J, et al. Endocrinology. 2016 Jun;157(6):2259-69. doi: 10.1210/en.2015-2027. Epub 2016 Apr 1. Endocrinology. 2016. PMID: 27035655 - Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1.
Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Nakahata Y, et al. Science. 2009 May 1;324(5927):654-7. doi: 10.1126/science.1170803. Epub 2009 Mar 12. Science. 2009. PMID: 19286518 Free PMC article. - The time of metabolism: NAD+, SIRT1, and the circadian clock.
Bellet MM, Orozco-Solis R, Sahar S, Eckel-Mahan K, Sassone-Corsi P. Bellet MM, et al. Cold Spring Harb Symp Quant Biol. 2011;76:31-8. doi: 10.1101/sqb.2011.76.010520. Epub 2011 Dec 16. Cold Spring Harb Symp Quant Biol. 2011. PMID: 22179986 Review. - Chromatin remodeling, metabolism and circadian clocks: the interplay of CLOCK and SIRT1.
Grimaldi B, Nakahata Y, Kaluzova M, Masubuchi S, Sassone-Corsi P. Grimaldi B, et al. Int J Biochem Cell Biol. 2009 Jan;41(1):81-6. doi: 10.1016/j.biocel.2008.08.035. Epub 2008 Sep 4. Int J Biochem Cell Biol. 2009. PMID: 18817890 Review.
Cited by
- Circadian Rhythm in Adipose Tissue: Novel Antioxidant Target for Metabolic and Cardiovascular Diseases.
Man AWC, Xia N, Li H. Man AWC, et al. Antioxidants (Basel). 2020 Oct 9;9(10):968. doi: 10.3390/antiox9100968. Antioxidants (Basel). 2020. PMID: 33050331 Free PMC article. Review. - The roles of gut microbiota and circadian rhythm in the cardiovascular protective effects of polyphenols.
Man AWC, Xia N, Daiber A, Li H. Man AWC, et al. Br J Pharmacol. 2020 Mar;177(6):1278-1293. doi: 10.1111/bph.14850. Epub 2019 Oct 31. Br J Pharmacol. 2020. PMID: 31465555 Free PMC article. Review. - P19 Cells as a Model for Studying the Circadian Clock in Stem Cells before and after Cell Differentiation.
Mashhour A, Al Mansour Z, Al Hallaj AS, Ali R, Trivilegio T, Boudjelal M. Mashhour A, et al. J Circadian Rhythms. 2018 May 18;16:6. doi: 10.5334/jcr.157. J Circadian Rhythms. 2018. PMID: 30210566 Free PMC article. - Influence of feeding time on daily rhythms of locomotor activity, clock genes, and epigenetic mechanisms in the liver and hypothalamus of the European sea bass (Dicentrarchus labrax).
Samorì E, Rodríguez I, Oliver JA, Sánchez-Vázquez FJ, López-Olmeda JF. Samorì E, et al. Fish Physiol Biochem. 2025 Feb 13;51(1):50. doi: 10.1007/s10695-025-01461-7. Fish Physiol Biochem. 2025. PMID: 39945981 Free PMC article. - SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging.
Chang HC, Guarente L. Chang HC, et al. Cell. 2013 Jun 20;153(7):1448-60. doi: 10.1016/j.cell.2013.05.027. Cell. 2013. PMID: 23791176 Free PMC article.
References
- Akhtar RA, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12(7):540–550. - PubMed
- Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. - PubMed
- Duffield GE, et al. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12(7):551–557. - PubMed
- Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases