Identifying transmission cycles at the human-animal interface: the role of animal reservoirs in maintaining gambiense human african trypanosomiasis - PubMed (original) (raw)
Identifying transmission cycles at the human-animal interface: the role of animal reservoirs in maintaining gambiense human african trypanosomiasis
Sebastian Funk et al. PLoS Comput Biol. 2013.
Abstract
Many infections can be transmitted between animals and humans. The epidemiological roles of different species can vary from important reservoirs to dead-end hosts. Here, we present a method to identify transmission cycles in different combinations of species from field data. We used this method to synthesise epidemiological and ecological data from Bipindi, Cameroon, a historical focus of gambiense Human African Trypanosomiasis (HAT, sleeping sickness), a disease that has often been considered to be maintained mainly by humans. We estimated the basic reproduction number [Formula: see text] of gambiense HAT in Bipindi and evaluated the potential for transmission in the absence of human cases. We found that under the assumption of random mixing between vectors and hosts, gambiense HAT could not be maintained in this focus without the contribution of animals. This result remains robust under extensive sensitivity analysis. When using the distributions of species among habitats to estimate the amount of mixing between those species, we found indications for an independent transmission cycle in wild animals. Stochastic simulation of the system confirmed that unless vectors moved between species very rarely, reintroduction would usually occur shortly after elimination of the infection from human populations. This suggests that elimination strategies may have to be reconsidered as targeting human cases alone would be insufficient for control, and reintroduction from animal reservoirs would remain a threat. Our approach is broadly applicable and could reveal animal reservoirs critical to the control of other infectious diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
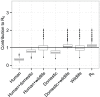
Figure 1. Contributions of species and species groups to under random mixing.
(a) The contributions of different species to  under the assumption of random mixing between vectors and hosts. (b) The contribution of different sets of species to
under the assumption of random mixing between vectors and hosts. (b) The contribution of different sets of species to  under the assumption of random mixing between vectors and hosts. In both plots, the y-axis shows the values of
under the assumption of random mixing between vectors and hosts. In both plots, the y-axis shows the values of  which would be found in a system of only the given (set of) species and vectors, the central line indicating the most likely value, upper and lower edges the interquartile range, the outer lines 1.5 times the interquartile range, and individual dots outlier results. The rightmost data point in (b) shows the estimate for
which would be found in a system of only the given (set of) species and vectors, the central line indicating the most likely value, upper and lower edges the interquartile range, the outer lines 1.5 times the interquartile range, and individual dots outlier results. The rightmost data point in (b) shows the estimate for  in the whole system (all species combined). Outliers for white-eyelid mangabeys with
in the whole system (all species combined). Outliers for white-eyelid mangabeys with  (0.1% of values) are not shown.
(0.1% of values) are not shown.
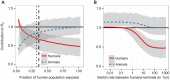
Figure 2. Human and animal contributions to under different model scenarios.
(a) The contribution of the human (red, solid) and animal (blue, dashed) populations to  as a function of the fraction of the population exposed to bites of the vector, shown here as effective population size
as a function of the fraction of the population exposed to bites of the vector, shown here as effective population size  . The vertical dashed line indicates the fraction of the population in the main endemic area , and the dotted line 90% of that population, a low estimate for screening efficacy . (b) The contribution of the human (red, solid) and animal (blue, dashed) populations to
. The vertical dashed line indicates the fraction of the population in the main endemic area , and the dotted line 90% of that population, a low estimate for screening efficacy . (b) The contribution of the human (red, solid) and animal (blue, dashed) populations to  as a function of the rate of host switching between a species, given in units of (number of switches)/year/fly. In both plots, the y-axis shows the values of
as a function of the rate of host switching between a species, given in units of (number of switches)/year/fly. In both plots, the y-axis shows the values of  which would be found in a system of only humans and the vector. The lines show the best estimate, and the light grey areas contain the smoothed (2.5%, 97.5%) quantile range, obtained from the binomial likelihood profiles and Latin hypercube sampling of parameter ranges (see Supporting Text S2).
which would be found in a system of only humans and the vector. The lines show the best estimate, and the light grey areas contain the smoothed (2.5%, 97.5%) quantile range, obtained from the binomial likelihood profiles and Latin hypercube sampling of parameter ranges (see Supporting Text S2).
Figure 3. Contributions of species groups to under habitat-specific mixing.
The contributions of different groups of species to  under the assumption of mixing proportional to habitat overlap of hosts. Hosts are grouped according to the habitats they can be found in, with random mixing within these groups and switching occurring at a third of the biting rate between the groups. The y-axis shows the values of
under the assumption of mixing proportional to habitat overlap of hosts. Hosts are grouped according to the habitats they can be found in, with random mixing within these groups and switching occurring at a third of the biting rate between the groups. The y-axis shows the values of  which would be found in a system of only the given set of species and vectors, the central line indicating the most likely value, upper and lower edges the interquartile range, the outer lines 1.5 times the interquartile range, and individual dots outlier results. The rightmost data point in shows the estimate for
which would be found in a system of only the given set of species and vectors, the central line indicating the most likely value, upper and lower edges the interquartile range, the outer lines 1.5 times the interquartile range, and individual dots outlier results. The rightmost data point in shows the estimate for  in the whole system (all species combined).
in the whole system (all species combined).
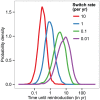
Figure 4. Reintroduction periods after elimination from the human and domestic population.
The probability distribution of reintroduction periods for different rates of host switching (given in units of (number of switches)/fly/year) between a human/domestic and a wild animal subsystem (with random mixing within each of these two subsystems), given in years. The values were obtained from  stochastic simulations, initialised with the prevalence in animal populations as measured in Bipindi, but with no infection present in humans, domestic animals, or human-associated vectors. Simulations were initialised with
stochastic simulations, initialised with the prevalence in animal populations as measured in Bipindi, but with no infection present in humans, domestic animals, or human-associated vectors. Simulations were initialised with  vectors, based on the number of around 2,000–3,000 flies captured in the area through entomological surveys lasting a few days . We considered reintroduction to have occurred once there were 2 cases in humans at any given time.
vectors, based on the number of around 2,000–3,000 flies captured in the area through entomological surveys lasting a few days . We considered reintroduction to have occurred once there were 2 cases in humans at any given time.
Similar articles
- Do Cryptic Reservoirs Threaten Gambiense-Sleeping Sickness Elimination?
Informal Expert Group on Gambiense HAT Reservoirs; Büscher P, Bart JM, Boelaert M, Bucheton B, Cecchi G, Chitnis N, Courtin D, Figueiredo LM, Franco JR, Grébaut P, Hasker E, Ilboudo H, Jamonneau V, Koffi M, Lejon V, MacLeod A, Masumu J, Matovu E, Mattioli R, Noyes H, Picado A, Rock KS, Rotureau B, Simo G, Thévenon S, Trindade S, Truc P, Van Reet N. Informal Expert Group on Gambiense HAT Reservoirs, et al. Trends Parasitol. 2018 Mar;34(3):197-207. doi: 10.1016/j.pt.2017.11.008. Epub 2018 Jan 23. Trends Parasitol. 2018. PMID: 29396200 Free PMC article. Review. - Molecular identification of Trypanosoma brucei gambiense in naturally infected pigs, dogs and small ruminants confirms domestic animals as potential reservoirs for sleeping sickness in Chad.
Vourchakbé J, Tiofack ZAA, Kante TS, Mpoame M, Simo G. Vourchakbé J, et al. Parasite. 2020;27:63. doi: 10.1051/parasite/2020061. Epub 2020 Nov 18. Parasite. 2020. PMID: 33206595 Free PMC article. - Diversity and spatial distribution of vectors and hosts of T. brucei gambiense in forest zones of, Southern Cameroon: epidemiological implications.
Massussi JA, Massussi JA, Mbida Mbida JA, Djieto-Lordon C, Njiokou F, Laveissière C, van der Ploeg JD. Massussi JA, et al. Acta Trop. 2010 Apr;114(1):44-8. doi: 10.1016/j.actatropica.2010.01.002. Epub 2010 Jan 11. Acta Trop. 2010. PMID: 20067756 - Wild fauna as a probable animal reservoir for Trypanosoma brucei gambiense in Cameroon.
Njiokou F, Laveissière C, Simo G, Nkinin S, Grébaut P, Cuny G, Herder S. Njiokou F, et al. Infect Genet Evol. 2006 Mar;6(2):147-53. doi: 10.1016/j.meegid.2005.04.003. Epub 2005 Oct 19. Infect Genet Evol. 2006. PMID: 16236560 - The natural progression of Gambiense sleeping sickness: what is the evidence?
Checchi F, Filipe JA, Barrett MP, Chandramohan D. Checchi F, et al. PLoS Negl Trop Dis. 2008;2(12):e303. doi: 10.1371/journal.pntd.0000303. Epub 2008 Dec 23. PLoS Negl Trop Dis. 2008. PMID: 19104656 Free PMC article. Review.
Cited by
- Diversity of trypanosomes in humans and cattle in the HAT foci Mandoul and Maro, Southern Chad-A matter of concern for zoonotic potential?
Ibrahim MAM, Weber JS, Ngomtcho SCH, Signaboubo D, Berger P, Hassane HM, Kelm S. Ibrahim MAM, et al. PLoS Negl Trop Dis. 2021 Jun 9;15(6):e0009323. doi: 10.1371/journal.pntd.0009323. eCollection 2021 Jun. PLoS Negl Trop Dis. 2021. PMID: 34106914 Free PMC article. - Molecular evidence of a Trypanosoma brucei gambiense sylvatic cycle in the human african trypanosomiasis foci of Equatorial Guinea.
Cordon-Obras C, Rodriguez YF, Fernandez-Martinez A, Cano J, Ndong-Mabale N, Ncogo-Ada P, Ndongo-Asumu P, Aparicio P, Navarro M, Benito A, Bart JM. Cordon-Obras C, et al. Front Microbiol. 2015 Jul 24;6:765. doi: 10.3389/fmicb.2015.00765. eCollection 2015. Front Microbiol. 2015. PMID: 26257727 Free PMC article. - Village-scale persistence and elimination of gambiense human African trypanosomiasis.
Davis CN, Rock KS, Mwamba Miaka E, Keeling MJ. Davis CN, et al. PLoS Negl Trop Dis. 2019 Oct 28;13(10):e0007838. doi: 10.1371/journal.pntd.0007838. eCollection 2019 Oct. PLoS Negl Trop Dis. 2019. PMID: 31658269 Free PMC article. - Update of transmission modelling and projections of gambiense human African trypanosomiasis in the Mandoul focus, Chad.
Rock KS, Huang CI, Crump RE, Bessell PR, Brown PE, Tirados I, Solano P, Antillon M, Picado A, Mbainda S, Darnas J, Crowley EH, Torr SJ, Peka M. Rock KS, et al. Infect Dis Poverty. 2022 Jan 24;11(1):11. doi: 10.1186/s40249-022-00934-8. Infect Dis Poverty. 2022. PMID: 35074016 Free PMC article. - The study of trypanosome species circulating in domestic animals in two human African trypanosomiasis foci of Côte d'Ivoire identifies pigs and cattle as potential reservoirs of Trypanosoma brucei gambiense.
N'Djetchi MK, Ilboudo H, Koffi M, Kaboré J, Kaboré JW, Kaba D, Courtin F, Coulibaly B, Fauret P, Kouakou L, Ravel S, Deborggraeve S, Solano P, De Meeûs T, Bucheton B, Jamonneau V. N'Djetchi MK, et al. PLoS Negl Trop Dis. 2017 Oct 18;11(10):e0005993. doi: 10.1371/journal.pntd.0005993. eCollection 2017 Oct. PLoS Negl Trop Dis. 2017. PMID: 29045405 Free PMC article.
References
- Mulligan HW, Potts WH (1970) The African trypanosomiases. Hoboken: Wiley. 950 pp.
- Hoare CA (1972) The trypanosomes of mammals. A zoological monograph. Oxford: Blackwell. 749 pp.
- Leak SGA (1999) Tsetse Biology and Ecology: Their Role in the Epidemiology and Control of Trypanosmosis. Wallingford: CABI. 448 pp.
- Maudlin I, Holmes PH, Miles MA (2004) The Trypanosomiases. Wallingford: CABI.
Publication types
MeSH terms
Grants and funding
SF was supported by EU FP7 funded integrated project EPIWORK (grant agreement number 231807). HN was supported by the JST PRESTO program. FC was supported by an AXA Research Fund Post-Doctoral Fellowship. This work was also assisted through participation in the Mathematical Modeling of Wildlife Zoonoses Investigative Workshop at the National Institute for Mathematical and Biological Synthesis, sponsored by the National Science Foundation, the U.S. Department of Homeland Security, and the U.S. Department of Agriculture through NSF Award #EF-0832858, with additional support from The University of Tennessee, Knoxville. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources