An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2 - PubMed (original) (raw)
An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2
Farida Korobova et al. Science. 2013.
Abstract
Mitochondrial fission is fundamentally important to cellular physiology. The dynamin-related protein Drp1 mediates fission, and interaction between mitochondrion and endoplasmic reticulum (ER) enhances fission. However, the mechanism for Drp1 recruitment to mitochondria is unclear, although previous results implicate actin involvement. Here, we found that actin polymerization through ER-localized inverted formin 2 (INF2) was required for efficient mitochondrial fission in mammalian cells. INF2 functioned upstream of Drp1. Actin filaments appeared to accumulate between mitochondria and INF2-enriched ER membranes at constriction sites. Thus, INF2-induced actin filaments may drive initial mitochondrial constriction, which allows Drp1-driven secondary constriction. Because INF2 mutations can lead to Charcot-Marie-Tooth disease, our results provide a potential cellular mechanism for this disease state.
Figures
Fig. 1
INF2 suppression increases mitochondrial length. (A) Maximum intensity projections of confocal images of MitoTracker-labeled U2OS cells treated with the indicated siRNAs. Scale bar, 20 μm. (B) Quantification of mitochondrial lengths. n = 157 to 531 mitochondria. Error bars, SEM.
Fig. 2
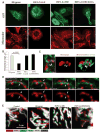
Constitutively active INF2-CAAX decreases mitochondrial length and dynamics. (A) Micrographs of U2OS cells transfected with GFP-fusions and labeled with MitoTracker. INF2 constructs are CAAX. Scale bar, 20 μm. (B) Quantifications of mitochondrial length. n = 158 to 537 mitochondria. Error bars, SEM. (C) Confocal micrographs of mitochondrion in close association with INF2-A149D. (Left) Maximum intensity projection of z stack. (Right) 3D reconstruction of MitoTracker alone or MitoTracker with GFP overlay. Arrowheads indicate constriction and/or fission sites. Scale bar, 2 μm. (D) Mitochondrial dynamics (mitochondria labeled using Mito Red) in U2OS transfected with GFP–INF2-A149D. Arrows indicate the fission event. Scale bar, 5 μm. (E) Overlays of mitochondrial dynamics time course in cells transfected with indicated constructs. Colors depict time points in the sequence, with white color indicating relative immobility.
Fig. 3
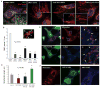
INF2 enhances Drp1-mitochondria association. (A) Confocal micrographs of U2OS cells labeled with an antibody against Drp1 (green) after indicated siRNA treatment. MitoTracker staining (red), and 4′,6-diamidino-2-phenylindole (DAPI) staining (blue). (Insets) Close-ups of peripheral cellular regions. Scale bar, 20 μm. (B) Quantification of Drp1 puncta per mitochondrial length, n = 40 to 68 mitochondria. Error bars, SEM. (C) Example of Drp1 puncta localization to mitochondria (arrowheads). (D) Effect of INF2-A149D expression (green) on Drp1 puncta. Mitochondria (red), Drp1 (blue). (Right) Close-ups of regions in INF2-A149D (1) and control cells (2). Scale bar, 20 μm. (E and F) Inhibition of Drp1 reduces the effect of INF2-A149D and makes mitochondria longer. (E) Quantification of mitochondrial length upon INF2-A149D expression and either Drp1 suppression or Drp1 K38E coexpression. n = 199 to 531 mitochondria. Error bars, SEM. (F) Confocal micrographs of U2OS cells coexpressing Mito Red (red), INF2-A149D (green), and Drp1 K38E (blue). Scale bar, 20 μm.
Fig. 4
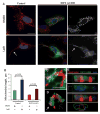
Actin filaments are required for INF2-mediated mitochondrial fission. (A) Control or INF2-A149D U2OS cells treated with dimethyl sulfoxide (DMSO) or 0.5 μM LatB for 60 min. Maximum intensity projections of six to nine z steps. Scale bar, 20 μm. Arrows indicate long mitochondria. (B) Quantification of mitochondrial lengths after DMSO or LatB. n = 53 to 110 mitochondria. Error bars, SD. (C to E) 3D reconstruction of mitochondrion (red) in INF2-A149D cell (green) labeled with Alexa660-phalloidin (blue). (C) Maximum intensity projection of nine z steps (D) Volume rendering of (C). (E) Same mitochondrion viewed along x axis [arrow in (D)], presented as 3D combinations. (Bottom) Outlines of stains to illustrate relative positions. Scale bar, 2 μm.
References
- Hoppins S, Nunnari J. Science. 2012;337:1052. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM069818/GM/NIGMS NIH HHS/United States
- GM069818/GM/NIGMS NIH HHS/United States
- R01 DK088826/DK/NIDDK NIH HHS/United States
- P30 CA023108/CA/NCI NIH HHS/United States
- DK88826/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous