Effector protein translocation by the Coxiella burnetii Dot/Icm type IV secretion system requires endocytic maturation of the pathogen-occupied vacuole - PubMed (original) (raw)
Effector protein translocation by the Coxiella burnetii Dot/Icm type IV secretion system requires endocytic maturation of the pathogen-occupied vacuole
Hayley J Newton et al. PLoS One. 2013.
Abstract
The human pathogen Coxiella burnetii encodes a type IV secretion system called Dot/Icm that is essential for intracellular replication. The Dot/Icm system delivers bacterial effector proteins into the host cytosol during infection. The effector proteins delivered by C. burnetii are predicted to have important functions during infection, but when these proteins are needed during infection has not been clearly defined. Here, we use a reporter system consisting of fusion proteins that have a β-lactamase enzyme (BlaM) fused to C. burnetii effector proteins to study protein translocation by the Dot/Icm system. Translocation of BlaM fused to the effector proteins CBU0077, CBU1823 and CBU1524 was not detected until 8-hours after infection of HeLa cells, which are permissive for C. burnetii replication. Translocation of these effector fusion proteins by the Dot/Icm system required acidification of the Coxiella-containing vacuole. Silencing of the host genes encoding the membrane transport regulators Rab5 or Rab7 interfered with effector translocation, which indicates that effectors are not translocated until bacteria traffic to a late endocytic compartment in the host cell. Similar requirements for effector translocation were discerned in bone marrow macrophages derived from C57BL/6 mice, which are primary cells that restrict the intracellular replication of C. burnetii. In addition to requiring endocytic maturation of the vacuole for Dot/Icm-mediated translocation of effectors, bacterial transcription was required for this process. Thus, translocation of effector proteins by the C. burnetii Dot/Icm system occurs after acidification of the CCV and maturation of this specialized organelle to a late endocytic compartment. This indicates that creation of the specialized vacuole in which C. burnetii replicates represents a two-stage process mediated initially by host factors that regulate endocytic maturation and then by bacterial effectors delivered into host cells after bacteria establish residency in a lysosome-derived organelle.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
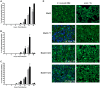
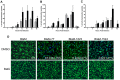
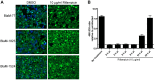
Figure 1. Time course of Dot/Icm dependent translocation.
C. burnetii NM and the icmL::Tn derivative expressing BlaM-77 (A), BlaM-1823 (B) or BlaM-1524 (C) from a plasmid were grown to stationary phase in ACCM-2, enumerated by qPCR, and used to infect HeLa cells at an MOI of 100 (grey bars) and 500 (black bars). At defined times post-infection the BlaM substrate CCF4-AM was added. Low magnification images for each sample were collected and used to calculate the percentage of cells that were translocation positive. This was determined by visual observation of a blue fluorescent emission at 460 nm when the cells were excited at 415 nm. At least 300 cells were quantified per well and each infection was performed in triplicate wells. These experiments were performed at least three independent times. Representative fluorescent micrographs of MOI 100 infections at 24 hours post-infection demonstrate the robust translocation of BlaM-77, BlaM-1823 and BlaM-1524, but not BlaM alone, when expressed by C. burnetii NM (D). Green HeLa cells (emission at 535 nm) represent cells loaded with uncleaved CCF4-AM and blue cells (emission at 460 nm) are indicative of CCF4-AM cleaved by translocated BlaM. The mean ± standard deviation percentage of cells that were translocation positive (blue) is displayed in the bottom right corner of each micrograph. No translocation was detected for C. burnetii expressing BlaM alone or for C. burnetii NM icmL::Tn expressing any of the reporter constructs.
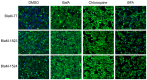
Figure 2. Acidification of the CCV is required for Dot/Icm dependent effector translocation.
Translocation assays were performed in the presence of chemical inhibitors to determine host requirements for translocation by the C. burnetii Dot/Icm secretion system. HeLa cells were infected with C. burnetii NM expressing BlaM, BlaM-77, BlaM-1823 or BlaM-1524 at a MOI of 100 and the infection was allowed to proceed for 24 h before translocation positive cells were quantified. At the time of infection, either BafA (100 nM), chloroquine (100 µM), BFA (1 µg/ml) or an equivalent volume of DMSO were also applied to the cells. DMSO and BFA treatments did not significantly alter the translocation efficiency of the BlaM reporters however no translocation was detected in the presence of BafA or chloroquine. Fluorescent micrographs are representative images of at least three independent experiments.
Figure 3. Translocation of Dot/Icm effector proteins is dependent on membrane transport of the vacuole to a late endocytic organelle.
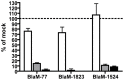
HeLa cells were transfected with pools of siRNA specific for Rab1 (white bars), Rab5 (grey bars), Rab7 (black bars) or mock transfected and incubated for three days before being infected with C. burnetii BlaM reporter strains at an MOI of 100 for 24 h. Translocation was visually quantified and is presented as the mean ± standard deviation relative to mock-transfected cells from a representative experiment.
Figure 4. Effector translocation in restrictive bone marrow macrophages.
Stationary phase C. burnetii NM expressing BlaM-77 (A), BlaM-1823 (B) or BlaM-1524 (C) were used to infect BMMs derived from C57BL/6 mice at an MOI of 100 (grey bars) and 500 (black bars). CCF4-AM was added at the time points shown and low magnification images were analyzed to calculate the percentage of cells that were translocation positive. At least 300 cells were quantified per well and each infection was performed in triplicate wells. These experiments were performed at least three independent times. Representative fluorescent micrographs of MOI 100 infections at 24 hours post-infection demonstrate translocation of BlaM-77, BlaM-1823 and BlaM-1524 (D). The mean ± standard deviation percentage of cells that were translocation positive (blue) is displayed in the bottom right corner of each micrograph. Additionally, the inclusion of 100 nM BafA at the time of infection led to no detectable translocation for any of the reporters (D). No translocation was detected for C. burnetii expressing BlaM alone or for C. burnetii NM icmL::Tn expressing any of the reporter constructs.
Figure 5. Effector translocation is dependent on C. burnetii transcription.
Inhibition of bacterial RNA synthesis through the addition of rifampicin blocks the capacity of C. burnetii to translocate BlaM-effector reporter proteins. No translocation of BlaM-77, BlaM-1823 or BlaM-1524 is detected after a 24 h infection in the presence of 10 μg/ml rifampicin (A). HeLa cells were infected with C. burnetii pBlaM-77 at an MOI of 100 and rifampicin was added at the time of infection, 2, 6, 8, 16 and 20 hours post-infection (h pi) before translocation was measured at 24 h pi (B). Translocation of the BlaM-CBU0077 fusion protein was determined by measuring the change in the 460 nm/535 nm fluorescence emission ratio resulting from cleavage of the CCF4-AM substrate (y-axis). Results represent the mean ± SD obtained from triplicate samples of a representative experiment.
Similar articles
- Coxiella burnetii effector proteins that localize to the parasitophorous vacuole membrane promote intracellular replication.
Larson CL, Beare PA, Voth DE, Howe D, Cockrell DC, Bastidas RJ, Valdivia RH, Heinzen RA. Larson CL, et al. Infect Immun. 2015 Feb;83(2):661-70. doi: 10.1128/IAI.02763-14. Epub 2014 Nov 24. Infect Immun. 2015. PMID: 25422265 Free PMC article. - Host pathways important for Coxiella burnetii infection revealed by genome-wide RNA interference screening.
McDonough JA, Newton HJ, Klum S, Swiss R, Agaisse H, Roy CR. McDonough JA, et al. mBio. 2013 Jan 29;4(1):e00606-12. doi: 10.1128/mBio.00606-12. mBio. 2013. PMID: 23362322 Free PMC article. - A screen of Coxiella burnetii mutants reveals important roles for Dot/Icm effectors and host autophagy in vacuole biogenesis.
Newton HJ, Kohler LJ, McDonough JA, Temoche-Diaz M, Crabill E, Hartland EL, Roy CR. Newton HJ, et al. PLoS Pathog. 2014 Jul 31;10(7):e1004286. doi: 10.1371/journal.ppat.1004286. eCollection 2014 Jul. PLoS Pathog. 2014. PMID: 25080348 Free PMC article. - Coxiella burnetii secretion systems.
McDonough JA, Newton HJ, Roy CR. McDonough JA, et al. Adv Exp Med Biol. 2012;984:171-97. doi: 10.1007/978-94-007-4315-1_9. Adv Exp Med Biol. 2012. PMID: 22711632 Review. - Coxiella type IV secretion and cellular microbiology.
Voth DE, Heinzen RA. Voth DE, et al. Curr Opin Microbiol. 2009 Feb;12(1):74-80. doi: 10.1016/j.mib.2008.11.005. Epub 2009 Jan 12. Curr Opin Microbiol. 2009. PMID: 19144560 Free PMC article. Review.
Cited by
- Murine Alveolar Macrophages Are Highly Susceptible to Replication of Coxiella burnetii Phase II In Vitro.
Fernandes TD, Cunha LD, Ribeiro JM, Massis LM, Lima-Junior DS, Newton HJ, Zamboni DS. Fernandes TD, et al. Infect Immun. 2016 Aug 19;84(9):2439-48. doi: 10.1128/IAI.00411-16. Print 2016 Sep. Infect Immun. 2016. PMID: 27297388 Free PMC article. - A Rab-centric perspective of bacterial pathogen-occupied vacuoles.
Sherwood RK, Roy CR. Sherwood RK, et al. Cell Host Microbe. 2013 Sep 11;14(3):256-68. doi: 10.1016/j.chom.2013.08.010. Cell Host Microbe. 2013. PMID: 24034612 Free PMC article. Review. - EirA Is a Novel Protein Essential for Intracellular Replication of Coxiella burnetii.
Kuba M, Neha N, Newton P, Lee YW, Bennett-Wood V, Hachani A, De Souza DP, Nijagal B, Dayalan S, Tull D, McConville MJ, Sansom FM, Newton HJ. Kuba M, et al. Infect Immun. 2020 May 20;88(6):e00913-19. doi: 10.1128/IAI.00913-19. Print 2020 May 20. Infect Immun. 2020. PMID: 32205404 Free PMC article. - Coxiella burnetii effector proteins that localize to the parasitophorous vacuole membrane promote intracellular replication.
Larson CL, Beare PA, Voth DE, Howe D, Cockrell DC, Bastidas RJ, Valdivia RH, Heinzen RA. Larson CL, et al. Infect Immun. 2015 Feb;83(2):661-70. doi: 10.1128/IAI.02763-14. Epub 2014 Nov 24. Infect Immun. 2015. PMID: 25422265 Free PMC article. - Caspase-8 activity mediates TNFα production and restricts Coxiella burnetii replication during murine macrophage infection.
Osbron CA, Lawson C, Hanna N, Koehler HS, Goodman AG. Osbron CA, et al. Infect Immun. 2024 Jul 11;92(7):e0005324. doi: 10.1128/iai.00053-24. Epub 2024 Jun 5. Infect Immun. 2024. PMID: 38837340 Free PMC article.
References
- Backert S, Meyer TF (2006) Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol 9: 207–217. - PubMed
- Hubber A, Roy CR (2010) Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 26: 261–283. - PubMed
- Segal G, Shuman HA (1999) Possible origin of the Legionella pneumophila virulence genes and their relation to Coxiella burnetii . Mol Microbiol 33: 669–670. - PubMed
- Berger KH, Isberg RR (1993) Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila . Mol Microbiol 7: 7–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54-AI057158/AI/NIAID NIH HHS/United States
- U54 AI057158/AI/NIAID NIH HHS/United States
- F32-AI0829272/AI/NIAID NIH HHS/United States
- R01 AI064559/AI/NIAID NIH HHS/United States
- R01-AI064559/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources