Silence, activate, poise and switch! Mechanisms of antigenic variation in Plasmodium falciparum - PubMed (original) (raw)
Review
Silence, activate, poise and switch! Mechanisms of antigenic variation in Plasmodium falciparum
Julien Guizetti et al. Cell Microbiol. 2013 May.
Free PMC article
Abstract
Phenotypic variation in genetically identical malaria parasites is an emerging topic. Although antigenic variation is only part of a more global parasite strategy to create adaptation through epigenetically controlled transcriptional variability, it is the central mechanism enabling immune evasion and promoting pathogenesis. The var gene family is the best-studied example in a wide range of clonally variant gene families in Plasmodium falciparum. It is unique in its strict selection of a single member for activation, a process termed monoallelic expression. The conceptual advances that have emerged from studying var genes show striking common epigenetic features with many other clonally variant gene families or even single-copy genes that show a variegated expression in parasite populations. However, major mechanistic questions, such as the existence of a potential expression site and the identity of transcription factors or genetic elements driving singular gene choice, are still unanswered. In this review we discuss the recent findings in the molecular processes essential for clonal variation, namely silencing, activation, poising and switching. Integrating findings about all clonally variant gene families and other mutually exclusive expression systems will hopefully drive mechanistic understanding of antigenic variation.
© 2013 Blackwell Publishing Ltd.
Figures
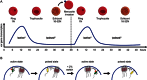
Fig. 1
Var gene activation and silencing throughout asexual blood stage development of P. falciparum. A. The single var gene is transcribed at the beginning of the asexual blood stage cycle (ring) soon after merozoites invasion of a red blood cells. Shortly before parasite DNA replication starts, var gene transcription ceases but it remains in a poised state (trophozoite and schizont) ready to be activated at the next blood stage cycle. B. All var genes, central as well as subtelomeric ones, are tethered to the nuclear periphery (blue) and silent var genes (red) cluster in repressive centers (grey) (Lopez-Rubio et al., 2009). A single active var gene (green) segregates away from repressive centres and forms a perinuclear expression site containing required transcription factors (orange). The transition from the active to the poised state (yellow) is still poorly defined and it is unknown whether positional memory establishes throughout late stages. However, recently a putative methyltransferase PfSet10 has been specifically associated with the poised var gene (Volz et al., 2012). Variable var gene switching rates have been measured in vitro (Horrocks et al., 2004), but it is unclear at which point in the cell cycle switching occurs.
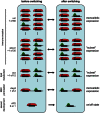
Fig. 2
Epigenetic switching in different clonally variant gene families of P. falciparum. Several gene families can be differentially expressed in genetically identical parasites (Rovira-Graells et al., 2012). The number of active genes (green) varies but silent genes are widely associated with epigenetic silencing mark H3K9me3 (red) (Lopez-Rubio et al., 2009). Combinatorial switching of the different gene families can result in a tremendous range of phenotypic variation within genetically identical populations. Gene families involved in immune evasion have different numbers, switching rates and strictness in gene counting. While var genes undergo strict monoallelic expression, multiple stevor genes can be transcribed in the same cell (Kaviratne et al., 2002). Gene families not involved in immune evasion can also undergo clonal variation. Here three examples with different ‘switching modes’ are shown. From the 12 members of the acs gene family, encoding for acyl-CoA synthetases 4 members have been shown to be clonally variant in blood stages (Rovira-Graells et al., 2012). The clag3 family, containing two members important for red blood cell permeation pathways, is the only other gene family besides var for which mutually exclusive expression has been shown (Cortes et al., 2007). A single member of the putative AP2 transcription family (PF3D7_1222600) is associated with H3K9me3. It is thought to be involved in inducing gametocytogenesis (Kafsack et al., MPM meeting 2012 Woods Hole). In this simple form epigenetically regulated activation at low frequency could constitute a developmental switch in the life cycle.
Similar articles
- CRISPR Interference of a Clonally Variant GC-Rich Noncoding RNA Family Leads to General Repression of var Genes in Plasmodium falciparum.
Barcons-Simon A, Cordon-Obras C, Guizetti J, Bryant JM, Scherf A. Barcons-Simon A, et al. mBio. 2020 Jan 21;11(1):e03054-19. doi: 10.1128/mBio.03054-19. mBio. 2020. PMID: 31964736 Free PMC article. - A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria.
Voss TS, Healer J, Marty AJ, Duffy MF, Thompson JK, Beeson JG, Reeder JC, Crabb BS, Cowman AF. Voss TS, et al. Nature. 2006 Feb 23;439(7079):1004-8. doi: 10.1038/nature04407. Epub 2005 Dec 28. Nature. 2006. PMID: 16382237 - A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria Parasite Plasmodium falciparum.
Ukaegbu UE, Zhang X, Heinberg AR, Wele M, Chen Q, Deitsch KW. Ukaegbu UE, et al. PLoS Genet. 2015 May 19;11(5):e1005234. doi: 10.1371/journal.pgen.1005234. eCollection 2015 May. PLoS Genet. 2015. PMID: 25993442 Free PMC article. - A greedy promoter controls malarial var-iations.
Scherf A. Scherf A. Cell. 2006 Jan 27;124(2):251-3. doi: 10.1016/j.cell.2006.01.004. Cell. 2006. PMID: 16439197 Review. - Antigenic variation in Plasmodium falciparum.
Scherf A, Lopez-Rubio JJ, Riviere L. Scherf A, et al. Annu Rev Microbiol. 2008;62:445-70. doi: 10.1146/annurev.micro.61.080706.093134. Annu Rev Microbiol. 2008. PMID: 18785843 Review.
Cited by
- Multiple Plasmodium falciparum Erythrocyte Membrane Protein 1 Variants per Genome Can Bind IgM via Its Fc Fragment Fcμ.
Jeppesen A, Ditlev SB, Soroka V, Stevenson L, Turner L, Dzikowski R, Hviid L, Barfod L. Jeppesen A, et al. Infect Immun. 2015 Oct;83(10):3972-81. doi: 10.1128/IAI.00337-15. Epub 2015 Jul 27. Infect Immun. 2015. PMID: 26216422 Free PMC article. - Post-translational protein modifications in malaria parasites.
Doerig C, Rayner JC, Scherf A, Tobin AB. Doerig C, et al. Nat Rev Microbiol. 2015 Mar;13(3):160-72. doi: 10.1038/nrmicro3402. Epub 2015 Feb 9. Nat Rev Microbiol. 2015. PMID: 25659318 Review. - Monoallelic expression and epigenetic inheritance sustained by a Trypanosoma brucei variant surface glycoprotein exclusion complex.
Faria J, Glover L, Hutchinson S, Boehm C, Field MC, Horn D. Faria J, et al. Nat Commun. 2019 Jul 9;10(1):3023. doi: 10.1038/s41467-019-10823-8. Nat Commun. 2019. PMID: 31289266 Free PMC article. - The conserved clag multigene family of malaria parasites: essential roles in host-pathogen interaction.
Gupta A, Thiruvengadam G, Desai SA. Gupta A, et al. Drug Resist Updat. 2015 Jan;18:47-54. doi: 10.1016/j.drup.2014.10.004. Epub 2014 Nov 3. Drug Resist Updat. 2015. PMID: 25467627 Free PMC article. Review. - A single-cell atlas of Plasmodium falciparum transmission through the mosquito.
Real E, Howick VM, Dahalan FA, Witmer K, Cudini J, Andradi-Brown C, Blight J, Davidson MS, Dogga SK, Reid AJ, Baum J, Lawniczak MKN. Real E, et al. Nat Commun. 2021 May 27;12(1):3196. doi: 10.1038/s41467-021-23434-z. Nat Commun. 2021. PMID: 34045457 Free PMC article.
References
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. - PubMed
- Bachmann A, Predehl S, May J, Harder S, Burchard GD, Gilberger TW, et al. Highly co-ordinated var gene expression and switching in clinical Plasmodium falciparum isolates from non-immune malaria patients. Cell Microbiol. 2011;9:1397–1409. - PubMed
- Balu B, Adams JH. Functional genomics of Plasmodium falciparum through transposon-mediated mutagenesis. Cell Microbiol. 2006;8:1529–1536. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources