Using whole-exome sequencing to identify inherited causes of autism - PubMed (original) (raw)
Case Reports
. 2013 Jan 23;77(2):259-73.
doi: 10.1016/j.neuron.2012.11.002.
Maria H Chahrour, Michael E Coulter, Sarn Jiralerspong, Kazuko Okamura-Ikeda, Bulent Ataman, Klaus Schmitz-Abe, David A Harmin, Mazhar Adli, Athar N Malik, Alissa M D'Gama, Elaine T Lim, Stephan J Sanders, Ganesh H Mochida, Jennifer N Partlow, Christine M Sunu, Jillian M Felie, Jacqueline Rodriguez, Ramzi H Nasir, Janice Ware, Robert M Joseph, R Sean Hill, Benjamin Y Kwan, Muna Al-Saffar, Nahit M Mukaddes, Asif Hashmi, Soher Balkhy, Generoso G Gascon, Fuki M Hisama, Elaine LeClair, Annapurna Poduri, Ozgur Oner, Samira Al-Saad, Sadika A Al-Awadi, Laila Bastaki, Tawfeg Ben-Omran, Ahmad S Teebi, Lihadh Al-Gazali, Valsamma Eapen, Christine R Stevens, Leonard Rappaport, Stacey B Gabriel, Kyriacos Markianos, Matthew W State, Michael E Greenberg, Hisaaki Taniguchi, Nancy E Braverman, Eric M Morrow, Christopher A Walsh
Affiliations
- PMID: 23352163
- PMCID: PMC3694430
- DOI: 10.1016/j.neuron.2012.11.002
Case Reports
Using whole-exome sequencing to identify inherited causes of autism
Timothy W Yu et al. Neuron. 2013.
Abstract
Despite significant heritability of autism spectrum disorders (ASDs), their extreme genetic heterogeneity has proven challenging for gene discovery. Studies of primarily simplex families have implicated de novo copy number changes and point mutations, but are not optimally designed to identify inherited risk alleles. We apply whole-exome sequencing (WES) to ASD families enriched for inherited causes due to consanguinity and find familial ASD associated with biallelic mutations in disease genes (AMT, PEX7, SYNE1, VPS13B, PAH, and POMGNT1). At least some of these genes show biallelic mutations in nonconsanguineous families as well. These mutations are often only partially disabling or present atypically, with patients lacking diagnostic features of the Mendelian disorders with which these genes are classically associated. Our study shows the utility of WES for identifying specific genetic conditions not clinically suspected and the importance of partial loss of gene function in ASDs.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
Figure 1
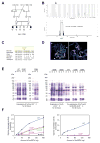
Identification of mutations in AMT in a family with ASD. (A) AU-1700, a Saudi family with three children affected by autism. Shaded symbols indicate affected individuals. The triangle represents a miscarriage. WES was performed on samples from individuals indicated with a star. Genotyping by Sanger sequencing in additional family members was performed where indicated (+: reference allele, −: alternate allele). (B) Mapping to a locus on chromosome 3. Genome-wide linkage plot (top) and maximum obtainable LOD score in the family across the interval (bottom). (C) Ile308 residue is highly conserved across species. (D) Mapping of the I308F missense mutation onto the human AMT crystal structure (PDB accession 1WSV). Left, overview showing I308 in domain 3 of AMT. Right, detail illustrating the hydrophobic pocket in which I308 resides. Neighboring hydrophobic residues are shown in white. The white brackets indicate the different domains: AMT domain 1 (folding); AMT domain 2 (catalytic); AMT domain 3 (capping). (E) I308F results in protein misfolding and aggregation. C-terminal 6xHis-tagged human AMT, AMT I308F, and AMT I308A were expressed in E. coli. Left panel, when overexpressed at 30°C by induction with 25 μM IPTG, wildtype AMT was fully soluble, but I308F and I308A segregated to inclusion bodies despite overall similar expression levels. W: whole cell extract, S: supernatant, P: pellet. Right panel, slower induction at 22°C for 44 hours resulted in partially soluble mutants. Near-wildtype solubility could be achieved by co-expression with GroEL and GroES. (F) AMT I308F results in partial loss-of-function of glycine cleavage and synthesis activity. Wildtype and mutant 6xHis-human AMT were expressed in E. coli, purified, and assayed for glycine cleavage and glycine synthesis activity. Relative to wildtype (blue traces), AMT I308F (red traces) demonstrates significant reduction of activity in glycine cleavage (left) and glycine synthesis (right) assays. R320H, N145I, and G269D are previously reported NKH-associated alleles (Okamura-Ikeda et al., 2005).
Figure 2
Identification of mutations in PEX7 in a family with ASD. (A) AU-3500, a family with three children affected with ASD. Shaded symbols indicate affected individuals. WES was performed on samples from individuals indicated with a star. Genotyping by Sanger sequencing in additional family members was performed where indicated (+: reference allele, −: alternate allele). (B) Mapping to a locus on chromosome 6. Genome-wide linkage plot (top) and maximum obtainable LOD score in the family across the interval (bottom). A homozygous missense mutation in the AU-3500 family disrupts a conserved tryptophan (p.W75C) (C) in the first WD40 repeat of PEX7, the receptor responsible for import of PTS2-containing proteins into the peroxisome (D), and an established cause of rhizomelic chondrodysplasia punctata. (E) PEX7 W75C and S25F, a previously characterized mutation from a patient with mild RCDP and ASD, result in partial loss-of-function in a peroxisomal import assay. In fibroblasts from patients with RCDP, PTS2-tagged mCherry accumulates in the cytoplasm due to lack of PEX7 transport activity (top). Transfection of wildtype PEX7 cDNA causes PTS2-mCherry to redistribute to small puncta, indicative of restoration of peroxisomal import (bottom). Transfection of mutant PEX7 W75C or S25F does not restore complete import, with a minority of cells showing partial import (middle). Quantification of defects in peroxisomal import mediated by ASD-associated PEX7 alleles, scored by visual assay. Error bars represent standard error (right). (F) Immunoblotting of whole cell lysates illustrates deficient maturation of PTS2-targeted proteins in a RCDP cell line transfected with ASD-associated PEX7 alleles. AGPS, PhyH, and thiolase are imported into the peroxisome and undergo cleavage into a smaller, mature peptide. p: preprotein, m: mature protein, Beta-tubulin: loading control. Quantification of processing defects by densitometry of transfected cell lysates (right).
Figure 3
Identification of mutations in SYNE1 in a family with ASD. (A) AU-1600, a family with four children affected with ASD. Shaded symbols indicate affected individuals. WES was performed on samples from individuals indicated with a star. Genotyping by Sanger sequencing in additional family members was performed where indicated (+: reference allele, −: alternate allele). (B) Mapping rules out the majority of the genome and points out to only two candidate loci, one on chromosome 6 and the other on chromosome 7. Genome-wide linkage plot (top) and maximum obtainable LOD score in the family across the interval (bottom). (C) SYNE1 is a complex locus implicated in neuropsychiatric disease. SYNE1 encompasses multiple alternative transcripts (Known Genes, UCSC Genome Browser), and mutations have been associated with a wide range of neurodevelopmental phenotypes. The location of the recessively inherited missense change in this study is denoted by the green star. Late truncation (open triangle) causes autosomal recessive arthrogyroposis multiplex congenita (Attali et al., 2009), while truncating mutations further upstream (closed triangles) cause autosomal recessive cerebellar ataxia (Gros-Louis et al., 2007). An intronic SNP (red square) is a putative bipolar disorder susceptibility locus (Sklar et al., 2011). A de novo missense change was previously reported in ASD (blue star) (O’Roak et al., 2011). (D) The homozygous missense mutation p.L3206M disrupts a highly conserved residue. (E) Human neuronal ChIPseq and RNAseq data demonstrate transcriptional complexity and activity-dependent regulation of the SYNE1 locus. ChIPseq with antibodies to H3K4Me3 (active promoters) and H3K27Ac (enhancers) demonstrates at least four active promoters (P1–P4) for SYNE1. P3 lies immediately upstream of the residue mutated in AU-1600 (p.L3206M) and corresponds to the active promoter site for BC039121. RNAseq of cultured human neurons was performed before (0h; 0 hours) and after (6h; 6 hours) depolarization with KCl. Depolarization induces upregulation of full-length SYNE1 and the BC039121 isoform compared to untreated neurons (also see Figure S3). The experiment was performed on five biological replicates. Representative tracks are shown. The red square represents a magnification of the region around BC039121. Sense (+; pink) and antisense (−; blue) transcripts are indicated.
Figure 4
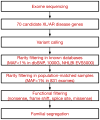
Schematic of the analytical pipeline. Tiered strategy for examining the cohort of ASD families for inherited mutations in known disease genes. Whole exome sequencing data was used to systematically survey 70 genes (listed in Table S2) known to be associated with autosomal recessive or X-linked neurodevelopmental illness and autistic features. Variants were filtered based on rarity (MAF<1%) in known databases and our internal, population-matched dataset (see Experimental Procedures for details), and predicted functional effect on the protein. All candidate variants were validated by Sanger sequencing and were required to segregate within the family. XL: X-linked, AR: autosomal recessive, MAF: minor allele frequency, 1000G: 1000 Genomes Project, NHLBI EVS5000: NHLBI Exome Sequencing Project.
Figure 5
Identification of a mutation in VPS13B, the Cohen syndrome gene in a patient with ASD. (A) AU-21100, a family with one child affected with ASD. Shaded symbol indicates affected individual. The triangle represents a miscarriage. WES was performed on samples from individuals indicated with a star. Genotyping by Sanger sequencing in additional family members was performed where indicated (+: reference allele, −: alternate allele). (B) Photographs of proband, demonstrating mild facial dysmorphisms consistent with Cohen syndrome: prominent nasal tip, protruding maxilla, and short philtrum. For additional clinical details, see Supplemental Text. (C) Head circumference for proband compared to published population normative data (Schienkiewitz et al., 2011). KiGGS: norms from German Health Interview and Examination Survey for Children and Adolescents, 2003–2006. Prader: previously published Swiss growth curves from 1989.
Comment in
- Rare inherited variation in autism: beginning to see the forest and a few trees.
Stein JL, Parikshak NN, Geschwind DH. Stein JL, et al. Neuron. 2013 Jan 23;77(2):209-11. doi: 10.1016/j.neuron.2013.01.010. Neuron. 2013. PMID: 23352155 Free PMC article.
Similar articles
- Whole Exome Sequencing Identifies Novel De Novo Variants Interacting with Six Gene Networks in Autism Spectrum Disorder.
Kim N, Kim KH, Lim WJ, Kim J, Kim SA, Yoo HJ. Kim N, et al. Genes (Basel). 2020 Dec 22;12(1):1. doi: 10.3390/genes12010001. Genes (Basel). 2020. PMID: 33374967 Free PMC article. - Mutations of ANK3 identified by exome sequencing are associated with autism susceptibility.
Bi C, Wu J, Jiang T, Liu Q, Cai W, Yu P, Cai T, Zhao M, Jiang YH, Sun ZS. Bi C, et al. Hum Mutat. 2012 Dec;33(12):1635-8. doi: 10.1002/humu.22174. Epub 2012 Aug 24. Hum Mutat. 2012. PMID: 22865819 - Identification of autism-related MECP2 mutations by whole-exome sequencing and functional validation.
Wen Z, Cheng TL, Li GZ, Sun SB, Yu SY, Zhang Y, Du YS, Qiu Z. Wen Z, et al. Mol Autism. 2017 Aug 3;8:43. doi: 10.1186/s13229-017-0157-5. eCollection 2017. Mol Autism. 2017. PMID: 28785396 Free PMC article. - The genetics of autism.
Muhle R, Trentacoste SV, Rapin I. Muhle R, et al. Pediatrics. 2004 May;113(5):e472-86. doi: 10.1542/peds.113.5.e472. Pediatrics. 2004. PMID: 15121991 Review. - Exome sequencing as a tool for Mendelian disease gene discovery.
Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Bamshad MJ, et al. Nat Rev Genet. 2011 Sep 27;12(11):745-55. doi: 10.1038/nrg3031. Nat Rev Genet. 2011. PMID: 21946919 Review.
Cited by
- Accessorizing and anchoring the LINC complex for multifunctionality.
Chang W, Worman HJ, Gundersen GG. Chang W, et al. J Cell Biol. 2015 Jan 5;208(1):11-22. doi: 10.1083/jcb.201409047. J Cell Biol. 2015. PMID: 25559183 Free PMC article. Review. - A germline mutation in SRRM2, a splicing factor gene, is implicated in papillary thyroid carcinoma predisposition.
Tomsic J, He H, Akagi K, Liyanarachchi S, Pan Q, Bertani B, Nagy R, Symer DE, Blencowe BJ, de la Chapelle A. Tomsic J, et al. Sci Rep. 2015 Jul 2;5:10566. doi: 10.1038/srep10566. Sci Rep. 2015. PMID: 26135620 Free PMC article. - Mammalian target of rapamycin pathway mutations cause hemimegalencephaly and focal cortical dysplasia.
D'Gama AM, Geng Y, Couto JA, Martin B, Boyle EA, LaCoursiere CM, Hossain A, Hatem NE, Barry BJ, Kwiatkowski DJ, Vinters HV, Barkovich AJ, Shendure J, Mathern GW, Walsh CA, Poduri A. D'Gama AM, et al. Ann Neurol. 2015 Apr;77(4):720-5. doi: 10.1002/ana.24357. Epub 2015 Feb 26. Ann Neurol. 2015. PMID: 25599672 Free PMC article. - Mechanism of human α3β GlyR modulation in inflammatory pain and 2, 6-DTBP interaction.
Wang W, Liu X. Wang W, et al. Res Sq [Preprint]. 2024 Aug 7:rs.3.rs-4402878. doi: 10.21203/rs.3.rs-4402878/v1. Res Sq. 2024. PMID: 39149480 Free PMC article. Updated. Preprint. - Somatic Mosaicism and Autism Spectrum Disorder.
D'Gama AM. D'Gama AM. Genes (Basel). 2021 Oct 26;12(11):1699. doi: 10.3390/genes12111699. Genes (Basel). 2021. PMID: 34828306 Free PMC article. Review.
References
- Applegarth DA, Toone JR. Nonketotic hyperglycinemia (glycine encephalopathy): laboratory diagnosis. Molecular Genetics and Metabolism. 2001;74:139–146. - PubMed
- Applegarth DA, Toone JR. Glycine encephalopathy (nonketotic hyperglycinaemia): review and update. Journal of inherited metabolic disease. 2004;27:417–422. - PubMed
- Attali R, Warwar N, Israel A, Gurt I, McNally E, Puckelwartz M, Glick B, Nevo Y, Ben-Neriah Z, Melki J. Mutation of SYNE-1, encoding an essential component of the nuclear lamina, is responsible for autosomal recessive arthrogryposis. Human Molecular Genetics. 2009;18:3462–3469. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 MH083565/MH/NIMH NIH HHS/United States
- T32 NS007473-12/NS/NINDS NIH HHS/United States
- T32 AG000222/AG/NIA NIH HHS/United States
- RC2 MH089952/MH/NIMH NIH HHS/United States
- T32 NS007484-08/NS/NINDS NIH HHS/United States
- P30 HD018655/HD/NICHD NIH HHS/United States
- 1RC2MH089952/MH/NIMH NIH HHS/United States
- T32 MH020017/MH/NIMH NIH HHS/United States
- R01 NS048276/NS/NINDS NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- T32GM07753/GM/NIGMS NIH HHS/United States
- K23 MH080954/MH/NIMH NIH HHS/United States
- K23NS069784/NS/NINDS NIH HHS/United States
- T32 NS007473/NS/NINDS NIH HHS/United States
- 1K23MH080954-01/MH/NIMH NIH HHS/United States
- K23 NS069784/NS/NINDS NIH HHS/United States
- T32 NS007484/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases