Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development - PubMed (original) (raw)
. 2013 Feb 11;24(3):310-23.
doi: 10.1016/j.devcel.2012.12.015. Epub 2013 Jan 24.
Achim Breiling, Thuc Le, Günter Raddatz, M Inmaculada Barrasa, Albert W Cheng, Qing Gao, Benjamin E Powell, Zhe Li, Mingjiang Xu, Kym F Faull, Frank Lyko, Rudolf Jaenisch
Affiliations
- PMID: 23352810
- PMCID: PMC3574201
- DOI: 10.1016/j.devcel.2012.12.015
Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development
Meelad M Dawlaty et al. Dev Cell. 2013.
Abstract
Tet enzymes (Tet1/2/3) convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) in various embryonic and adult tissues. Mice mutant for either Tet1 or Tet2 are viable, raising the question of whether these enzymes have overlapping roles in development. Here we have generated Tet1 and Tet2 double-knockout (DKO) embryonic stem cells (ESCs) and mice. DKO ESCs remained pluripotent but were depleted of 5hmC and caused developmental defects in chimeric embryos. While a fraction of double-mutant embryos exhibited midgestation abnormalities with perinatal lethality, viable and overtly normal Tet1/Tet2-deficient mice were also obtained. DKO mice had reduced 5hmC and increased 5mC levels and abnormal methylation at various imprinted loci. Nevertheless, animals of both sexes were fertile, with females having smaller ovaries and reduced fertility. Our data show that loss of both enzymes is compatible with development but promotes hypermethylation and compromises imprinting. The data also suggest a significant contribution of Tet3 to hydroxylation of 5mC during development.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
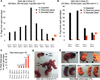
Figure 1. Tet1 Tet2 double knockout (DKO) ESCs are depleted of 5hmC and express pluripotency markers
(A) Schematic of crosses to generate DKO ESCs. (B) Summary of derived ESCs and Mendelian frequency of DKO ESC. (C) RTqPCR for Tet1, Tet2 and Tet3 ESCs of indicated genotypes. Notice that DKO ESCs are depleted of Tet1 and Tet2 mRNA. Data are normalized to Gapdh. (D) Bright field images of ESCs of indicated genotypes. (E) Immunostainning for pluripotency markers Oct4 and Nanog in ESCs. (F) Relative mRNA levels of Esrrb, Nanog and Oct4 in ESCs. Data are normalized to Gapdh. (G) WT and DKO ESCs 5-day cell proliferation count. (H & I) Quantification of 5hmC and 5mC in ESCs of indicated genotypes by mass spectrometry (LC-MS/MS-MRM). Notice in DKO ESCs 5hmC levels are below detection level. (J) Heat map of differentially expressed genes across a panel of wild type, single knockouts and DKOs ESCs. Genes differentially expressed in at least one of the comparisons are displayed. Intensity values are log2 transformed and genes are centered by mean intensity across all arrays. A total of 501 genes are deregulated in DKO ESCs. (K) Pie charts categorizing deregulated genes into sub categories of CpG rich genes (left), TSS 5hmC positive genes (center) and H3K4me and H3K27me bivalent genes (right) in ESCs. Asterisk indicates enriched and statistically significant. (L) Gene ontology analysis of 501 differentially expressed genes in DKO ESCs showing that most GO terms are implicated in development. For all panels m=male cell line and f=female cell line. Error bars indicate s.d. See also Figure S1 and Table S2.
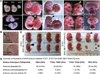
Figure 2. Double knockout ESCs remain pluripotent in teratoma and chimera assays
(A) Representative images of each germ layer from H&E stained sections of teratomas generated from ESCs of indicated genotypes. (B) High magnification images of H&E stained teratoma sections showing hemorrhage and trophoblast-like cells (arrowheads) in tumors derived from DKO and Tet1 deficient ESCs. (C) E14.5 chimeric embryos generated by injecting FUW-td-Tomato transduced ESCs of indicated genotypes into wild type blastocysts. Notice exencephaly and weaker td-tomato signal in DKO ESCs chimeras. (D) Summary table for all chimera injections and defects of DKO chimeric embryos.
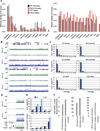
Figure 3. Partially penetrant perinatal lethality of Tet1/Tet2 deficient mice
(A &B) Charts summarizing percent number of pups for each genotype from indicated crosses. Note the increased number of neonatally dead pups with DKO genotype. Dead and alive pups add up close to the expected Mendelian ratio. Average litter sizes shown on top of the charts are calculated based on alive pups. (C) Total dead pups in the study (n=42) are classified based on their genotype. Notice that majority of dead pups are DKO or T1KOT2Het. (D) Gross images of P1 neonates. (E) Representative Images of neonatally lethal pups. See also Figure S2.
Figure 4. Defects in midgestation double knockout embryos
(A) Representative images of mid-gestation E10.5 to E15.5 embryos of indicated genotypes. Notice the presence of normal, smaller size and exencephalic DKO embryos. Arrowheads point to exencephaly/neural tube defects. All embryos are from crosses of DKO male × Dhet female. (B) H&E staining for histological analysis of exencephaly. (C) Images, genotypes and weights of E18.5 and E19.5 litters from the indicated crosses. Arrowheads point to exencephaly/neural tube defects. (D) Summary of genotypes, phenotypes and Mendelian frequency of all mid and late gestation embryos.
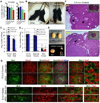
Figure 5. Fertility assessment of viable adult double knockout mice
(A&B) Average weight and gross images of DKO mice. Asterisks indicate p<0.05. (C) Average number of pups per litter from crosses of [DKO male × WT Females] or [Control WT males × WT females]. (D) Average number of pups per litter from crosses of [WT male × Control WT female] or [WT male × Dhet female] or [WT male × DKO female]. (E) Gross images of testis and ovaries. Notice the smaller size of ovaries. (F) H&E stained lateral sections of ovaries. Insets are low magnification images to highlight differences in size of ovaries. Notice the reduced presence of mature follicles and eggs (black arrows) and secondary follicles (white arrows) in DKO ovaries. (G) E13.5 gonad sections stained with anti-Mvh, anti 5hmC and anti 5mC antibodies and Dapi. Notice the lack of 5hmC staining in DKO germ cells. See also Figure S5.
Figure 6. Hypermethylation and reduced 5hmC levels in Tet1/Tet2 deficient embryos and adult mice
(A) Quantification of 5hmC and 5mC in various tissues of adult 2.5-month-old male and female mice by mass spectrometry. (B) hMeDIP-seq (green) and MeDIP-seq (blue) profiles of two 20 MB regions from mouse chromosome 4 (upper panel) and 19 (lower panel). MeDIP was performed on genomic DNA extracted from WT and DKO tails of neonates (pup#7 and #15, respectively). hMeDIP was performed on genomic DNA extracted from WT and DKO adult cerebrum. Enrichments are indicated as normalized read counts. UCSC transcription units are indicated on top in dark blue. Genomic features are viewed as custom tracks in the UCSC genome browser. (C) Quantitative analysis of MeDIP results. Normalized 5mC read counts for each sample were determined for all bases and then distributed into bins with increasing read counts (1–10). The resulting profiles from DKO samples show a consistent increase in the number of bases that are covered by a single read, representing low-level methylation. At the same time, the number of bases with higher coverage becomes decreased in DKO samples. Samples analysed were from WT and DKO neonates (MeDIP performed on DNA from tail/hind leg), embryos (MeDIP performed on DNA from total brain) and adults (MeDIP performed on DNA from cerebrum). (D) hMeDIP-seq and MeDIP-seq profiles (as in B) of the flanks of a CpG-rich repeat region on chromosome 6 (chr6:47,563,923-47,636,951). Genomic features are shown as custom tracks in the UCSC genome browser. (E) Quantitative MeDIP analysis of repetitive elements in WT (dark blue) and DKO (light blue) neonates. The upper panel shows normalized 5mC read counts for several classes of repetitive elements, as indicated. The lower panel shows the enrichment of repetitive element sequencing reads relative to the genome-wide average (indicated by the red line). Abbreviations indicate Long Interspersed Nuclear Elements (LINEs), Short Interspersed Nuclear Elements (SINEs), Long Terminal Repeats (LTRs), simple repeats (simp.) and satellite repeats (sat.). (F) Relative expression of Tet3 in various embryos and adult tissues. Data are normalized to Gapdh. For all panels error bars indicate s.d. See also Figure S3–S4.
Figure 7. Tet1/Tet2 deficiency partially compromises imprinting
(A, B, D, E) Heat map representation of bisulfite sequencing analysis of imprinted genes in selected samples corresponding to table S1. Within each heat map columns represent each CpG position in the sequenced ICR. Rows represent individual clones sequenced. Total of 16 to 20 clones were analyzed for each sample. Black box= methylated, white box=unmethylated and orange or grey box=undetermined due to poor sequence quality. Values on top of each map are % methylated CpG sites. Samples with aberrant (more than 65% methylation) are shown in red. Control samples are shown in blue. Detailed description of the entire analysis and all samples analyzed can be found in Table S1. (C) RTqPCR for Mest and Peg3 in selected E13.5 embryos confirming loss of expression of these genes in embryos with increased methylation shown in A and B. See also Table S1.
Similar articles
- Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development.
Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, Jaenisch R. Dawlaty MM, et al. Cell Stem Cell. 2011 Aug 5;9(2):166-75. doi: 10.1016/j.stem.2011.07.010. Cell Stem Cell. 2011. PMID: 21816367 Free PMC article. - Distinct and overlapping control of 5-methylcytosine and 5-hydroxymethylcytosine by the TET proteins in human cancer cells.
Putiri EL, Tiedemann RL, Thompson JJ, Liu C, Ho T, Choi JH, Robertson KD. Putiri EL, et al. Genome Biol. 2014 Jun 23;15(6):R81. doi: 10.1186/gb-2014-15-6-r81. Genome Biol. 2014. PMID: 24958354 Free PMC article. - Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification.
Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Ito S, et al. Nature. 2010 Aug 26;466(7310):1129-33. doi: 10.1038/nature09303. Nature. 2010. PMID: 20639862 Free PMC article. - Role of ten-eleven translocation proteins and 5-hydroxymethylcytosine in hepatocellular carcinoma.
Wang P, Yan Y, Yu W, Zhang H. Wang P, et al. Cell Prolif. 2019 Jul;52(4):e12626. doi: 10.1111/cpr.12626. Epub 2019 Apr 29. Cell Prolif. 2019. PMID: 31033072 Free PMC article. Review. - Tet family of 5-methylcytosine dioxygenases in mammalian development.
Zhao H, Chen T. Zhao H, et al. J Hum Genet. 2013 Jul;58(7):421-7. doi: 10.1038/jhg.2013.63. Epub 2013 May 30. J Hum Genet. 2013. PMID: 23719188 Free PMC article. Review.
Cited by
- The landscape of DNA methylation amid a perfect storm of autism aetiologies.
Vogel Ciernia A, LaSalle J. Vogel Ciernia A, et al. Nat Rev Neurosci. 2016 Jul;17(7):411-23. doi: 10.1038/nrn.2016.41. Epub 2016 May 6. Nat Rev Neurosci. 2016. PMID: 27150399 Free PMC article. Review. - Function and information content of DNA methylation.
Schübeler D. Schübeler D. Nature. 2015 Jan 15;517(7534):321-6. doi: 10.1038/nature14192. Nature. 2015. PMID: 25592537 Review. - TETs compete with DNMT3 activity in pluripotent cells at thousands of methylated somatic enhancers.
Charlton J, Jung EJ, Mattei AL, Bailly N, Liao J, Martin EJ, Giesselmann P, Brändl B, Stamenova EK, Müller FJ, Kiskinis E, Gnirke A, Smith ZD, Meissner A. Charlton J, et al. Nat Genet. 2020 Aug;52(8):819-827. doi: 10.1038/s41588-020-0639-9. Epub 2020 Jun 8. Nat Genet. 2020. PMID: 32514123 Free PMC article. - Epigenetic dysregulation of TET2 in human glioblastoma.
García MG, Carella A, Urdinguio RG, Bayón GF, Lopez V, Tejedor JR, Sierra MI, García-Toraño E, Santamarina P, Perez RF, Mangas C, Astudillo A, Corte-Torres MD, Sáenz-de-Santa-María I, Chiara MD, Fernández AF, Fraga MF. García MG, et al. Oncotarget. 2018 May 25;9(40):25922-25934. doi: 10.18632/oncotarget.25406. eCollection 2018 May 25. Oncotarget. 2018. PMID: 29899831 Free PMC article. - 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation.
Hon GC, Song CX, Du T, Jin F, Selvaraj S, Lee AY, Yen CA, Ye Z, Mao SQ, Wang BA, Kuan S, Edsall LE, Zhao BS, Xu GL, He C, Ren B. Hon GC, et al. Mol Cell. 2014 Oct 23;56(2):286-297. doi: 10.1016/j.molcel.2014.08.026. Epub 2014 Sep 25. Mol Cell. 2014. PMID: 25263596 Free PMC article.
References
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell. 2006;125:315–326. - PubMed
- Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2011:1–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5-RO1-HDO45022/PHS HHS/United States
- 5-R37-CA084198/CA/NCI NIH HHS/United States
- R37 HD045022/HD/NICHD NIH HHS/United States
- R37 CA084198/CA/NCI NIH HHS/United States
- R01 HL112294/HL/NHLBI NIH HHS/United States
- R01 HD045022/HD/NICHD NIH HHS/United States
- R01 CA084198/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases