Deregulation of Wnt/β-catenin signaling through genetic or epigenetic alterations in human neuroendocrine tumors - PubMed (original) (raw)
doi: 10.1093/carcin/bgt018. Epub 2013 Jan 25.
Jing Li, Eun Ryoung Jang, Pat Gulhati, Piotr G Rychahou, Dana L Napier, Chi Wang, Heidi L Weiss, Eun Y Lee, Lowell Anthony, Courtney M Townsend Jr, Chunming Liu, B Mark Evers
Affiliations
- PMID: 23354304
- PMCID: PMC3643417
- DOI: 10.1093/carcin/bgt018
Deregulation of Wnt/β-catenin signaling through genetic or epigenetic alterations in human neuroendocrine tumors
Ji Tae Kim et al. Carcinogenesis. 2013 May.
Abstract
Carcinoid tumors are rare neuroendocrine tumors (NETs) that are increasing in incidence. Mutation and altered expression of Wnt/β-catenin signaling components have been described in many tumors but have not been well-studied in NETs. Here, we observed accumulation of β-catenin in the cytoplasm and/or nucleus in 25% of clinical NET tissues. By mutational analysis, the mutations of β-catenin (I35S) and APC (E1317Q, T1493T) were identified in NET cells and the tissues. Expression of representative Wnt inhibitors was absent or markedly decreased in BON, a human pancreatic carcinoid cell line; treatment with 5-aza-2'-deoxycytidine (5-aza-CdR) increased expression levels of the Wnt inhibitors. Methylation analyses demonstrated that CpG islands of SFRP-1 and Axin-2 were methylated, whereas the promoters of DKK-1, DKK-3 and WIF-1 were unmethylated in four NET cells. Aberrant methylation of SFRP-1 was particularly observed in most of clinical NET tissues. In addition, the repression of these unmethylated genes was associated with histone H3 lysine 9 dimethylation (H3K9me2) in BON cells. Together, 5-aza-CdR treatment inhibited cell proliferation and decreased the protein levels of H3K9me2 and G9a. Moreover, a novel G9a inhibitor, UNC0638, suppressed BON cell proliferation through inhibition of Wnt/β-catenin pathway. Overexpression of the inhibitory genes, particularly SFRP-1 and WIF-1 in BON cells, resulted in suppression of anchorage-independent growth and inhibition of tumor growth in mice. Our findings suggest that aberrant Wnt/β-catenin signaling, through either mutations or epigenetic silencing of Wnt antagonists, contributes to the pathogenesis and growth of NETs and have important clinical implications for the prognosis and treatment of NETs.
Figures
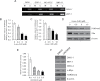
Fig. 1.
Analysis of β-catenin localization in clinical NET samples and direct DNA sequence analysis of β-catenin and APC in NET cell lines and clinical specimens. (A) Immunohistochemical analysis of β-catenin in clinical NET samples. Membranous staining of β-catenin without cytoplasmic and nuclear staining in a small bowel carcinoid tumor (upper). Cytoplasmic and membranous staining of β-catenin without nuclear staining in another small bowel carcinoid tumor (middle). Strong nuclear and cytoplasmic staining of β-catenin in a thymus carcinoid tumor (bottom). (B) I35S mutation in β-catenin (ATC to AGC substitution in nucleotide) in a thymus carcinoid tissue. (C) E1317Q mutation in APC (GAA to CAA substitution in nucleotide) in BON cells (upper). T1493T silent mutation in APC (ACG to ACA substitution in nucleotide) in BON cells (bottom). Asterisk indicates base substitution sites. (D) Summary of β-catenin and APC mutations in NET cell lines and clinical samples.
Fig. 2.
Expression analysis of Wnt/β-catenin inhibitor genes in BON cells after 5-aza-CdR treatment. (A) RT–PCR analysis of SFRP-1, Axin-2, DKK-1, DKK-3, WIF-1 and β-actin expression in BON cells with or without 5-aza-CdR treatment. Cells were treated with varying concentrations of 5-aza-CdR for 96h with media changed every 24h. Total RNA was isolated from BON cells treated with 0 [dimethyl sulfoxide (DMSO)], 0.5 or 5 μM 5-aza-CdR and cDNA was synthesized from 1 µg of total RNA. The reaction was performed using HotStarTaq DNA Polymerase (Qiagen) and the primers listed in
Supplementary Table 1
, available at Carcinogenesis Online. The PCR products were visualized by 2% agarose gel. (B) qRT–PCR analysis confirmed that treatment with 5-aza-CdR induced the restoration of the Wnt/β-catenin pathway inhibitor genes in BON cells. The reaction was performed using iCycler with the iQ SYBR-green Supermix (Bio-Rad), cDNA and the same primers. Each gene expression was assessed by evaluating threshold cycle (Ct) values. The relative amount of mRNA expression was calculated by the comparative ΔΔCt method. Columns represent triplicate data points; error bars, SD; *P < 0.05 versus DMSO treatment; β-actin was used as internal control in both analyses.
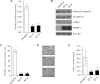
Fig. 3.
DNA methylation analyses of the 5′ region of the SFRP-1 and Axin-2 in NET cells and tissues. (A) MSP analysis of CpG islands of SFRP-1 and Axin-2 with respective two primer pairs (MSP-A and -B) and primers specific for the methylated (M) and unmethylated (U) DNA in four NET cells. NS represents non-specific bands or primer dimers for PCR products. (B) Bisulfite genomic sequencing analysis of two 5′ regions of SFRP-1 in BON and QGP-1 cells. (C) Bisulfite genomic sequencing analysis of Axin-2 CpG islands in BON and QGP-1 cells. Each row of circles represents the DNA sequence of an individual clone; closed and open circles indicate methylated and unmethylated CpG sites, respectively. (D) MSP analysis of the upstream region of the SFRP-1 and Axin-2 with the same primers described above in clinical NET samples (patient numbers).
Fig. 4.
Repression of DKK-1 and WIF-1 is associated with the level of H3K9me2 in BON cells. (A) MSP analysis with primers specific for the methylated (M) and unmethylated (U) DKK-1 and WIF-1 CpG islands in four NET cell lines. ChIP analysis demonstrated reduction of H3K9me2 within the DKK-1 (B) and WIF-1 (C) promoter in BON cells following 5-aza-CdR treatment for 96h. The amount of recruitment of H3K9me2 to the DKK-1 and WIF-1 promoter was quantified by real-time PCR and plotted as the H3K9me2 binding ratio of DKK-1 or WIF-1 to β-globin and then expressed relative to DMSO-treated BON cells. Columns represent triplicate data points; error bars, SD; *P < 0.05 versus DMSO treatment. (D) BON cells were treated with 5-aza-CdR for 96h and protein extracts were analyzed by western blotting with the indicated antibodies; β-actin was used as an internal control for protein loading. (E) Relative luciferase activity obtained using a TOPFlash and, as a control, a Renilla luciferase reporter in BON cells treated with indicated concentrations of UNC0638. Columns represent triplicate data points; error bars, SD; *P < 0.05 versus DMSO treatment. (F) Western blotting analysis of UNC0638 effects on expression of silenced proteins and its related proteins. BON cells were treated with 0 or 2.5 μM UNC0638 for 1 day and protein extracts were analyzed with the indicated antibodies.
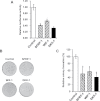
Fig. 5.
Tumor suppressive effects of Wnt inhibitor genes in BON cells. (A) Relative luciferase activity obtained using a TOPFlash and Renilla luciferase reporter in BON cells transiently transfected with control (empty vector) or the indicated Wnt inhibitor genes. Columns represent triplicate data points; error bars, SD; *P < 0.05 versus control. (B) Representative results from a colony formation assay carried out in BON cells transfected with control (empty vector) or the respective Wnt inhibitor gene constructs. (C) The relative number of colonies compared with the control; error bars, SD; *P < 0.05 versus control from triplicate analyses.
Fig. 6.
Tumor suppressive properties of SFRP-1 in BON cells. (A) Relative luciferase activity obtained using a TOPFlash and a Renilla in SFRP-1 BON cell clones (S1-2 and S1-4). Columns represent triplicate data points; error bars, SD; *P < 0.05 versus control (empty vector). (B) Western blot analysis showing expression of Wnt/β-catenin pathway-related proteins regulated by SFRP-1. Overexpression of SFRP-1 decreases the level of active-β-catenin and c-Myc, a target of Wnt/β-catenin signaling pathway. (C) The relative number of colonies compared with the control in soft agar assay. Colony formation of the BON cell clones was assessed over a period of 4 weeks. Columns represent triplicate data points; error bars, SD; *P < 0.05 versus control. (D) Representative results from a soft agar assay carried out in two SFRP-1 and control BON cell clones. (E) Overexpression of SFRP-1 suppresses growth of BON cell xenografts in vivo. Athymic nude mice were inoculated subcutaneously with the stable BON cell clones. Five mice were used in each group, and the cells were inoculated at one site in the flank of each mouse. Size of the tumors was measured after 3 weeks. Each value represents mean ± SD of tumor volume (mm3) (*P < 0.05 versus control).
Similar articles
- WNT pathway in oral cancer: epigenetic inactivation of WNT-inhibitors.
Pannone G, Bufo P, Santoro A, Franco R, Aquino G, Longo F, Botti G, Serpico R, Cafarelli B, Abbruzzese A, Caraglia M, Papagerakis S, Lo Muzio L. Pannone G, et al. Oncol Rep. 2010 Oct;24(4):1035-41. doi: 10.3892/or.2010.1035. Oncol Rep. 2010. PMID: 20811686 - Epigenetic inactivation of the Wnt antagonist DICKKOPF-1 (DKK-1) gene in human colorectal cancer.
Aguilera O, Fraga MF, Ballestar E, Paz MF, Herranz M, Espada J, García JM, Muñoz A, Esteller M, González-Sancho JM. Aguilera O, et al. Oncogene. 2006 Jul 6;25(29):4116-21. doi: 10.1038/sj.onc.1209439. Epub 2006 Feb 20. Oncogene. 2006. PMID: 16491118 - Extracellular activation of Wnt signaling through epigenetic dysregulation of Wnt inhibitory factor-1 (Wif-1) is associated with pathogenesis of adrenocortical tumor.
Mitsui Y, Yasumoto H, Nagami T, Hiraki M, Arichi N, Ishikawa N, Araki A, Maruyama R, Tanaka Y, Dahiya R, Shiina H. Mitsui Y, et al. Oncotarget. 2014 Apr 30;5(8):2198-207. doi: 10.18632/oncotarget.1889. Oncotarget. 2014. PMID: 24755523 Free PMC article. - The many ways of Wnt in cancer.
Polakis P. Polakis P. Curr Opin Genet Dev. 2007 Feb;17(1):45-51. doi: 10.1016/j.gde.2006.12.007. Curr Opin Genet Dev. 2007. PMID: 17208432 Review. - Epigenetic alterations of the Wnt/beta-catenin pathway in human disease.
Aguilera O, Muñoz A, Esteller M, Fraga MF. Aguilera O, et al. Endocr Metab Immune Disord Drug Targets. 2007 Mar;7(1):13-21. doi: 10.2174/187153007780059450. Endocr Metab Immune Disord Drug Targets. 2007. PMID: 17346200 Review.
Cited by
- Pancreatic Neuroendocrine Tumors: Molecular Mechanisms and Therapeutic Targets.
Maharjan CK, Ear PH, Tran CG, Howe JR, Chandrasekharan C, Quelle DE. Maharjan CK, et al. Cancers (Basel). 2021 Oct 12;13(20):5117. doi: 10.3390/cancers13205117. Cancers (Basel). 2021. PMID: 34680266 Free PMC article. Review. - A systems medicine approach for finding target proteins affecting treatment outcomes in patients with non-Hodgkin lymphoma.
Ajorloo F, Vaezi M, Saadat A, Safaee SR, Gharib B, Ghanei M, Siadat SD, Vaziri F, Fateh A, Pazhouhandeh M, Vaziri B, Moazemi R, Mahboudi F, Rahimi Jamnani F. Ajorloo F, et al. PLoS One. 2017 Sep 11;12(9):e0183969. doi: 10.1371/journal.pone.0183969. eCollection 2017. PLoS One. 2017. PMID: 28892521 Free PMC article. - NETs: organ-related epigenetic derangements and potential clinical applications.
Cives M, Simone V, Rizzo FM, Silvestris F. Cives M, et al. Oncotarget. 2016 Aug 30;7(35):57414-57429. doi: 10.18632/oncotarget.10598. Oncotarget. 2016. PMID: 27418145 Free PMC article. Review. - Neurotensin, a novel target of Wnt/β-catenin pathway, promotes growth of neuroendocrine tumor cells.
Kim JT, Liu C, Zaytseva YY, Weiss HL, Townsend CM Jr, Evers BM. Kim JT, et al. Int J Cancer. 2015 Mar 15;136(6):1475-81. doi: 10.1002/ijc.29123. Epub 2014 Aug 14. Int J Cancer. 2015. PMID: 25098665 Free PMC article. - From microbiota toward gastro-enteropancreatic neuroendocrine neoplasms: Are we on the highway to hell?
Vitale G, Dicitore A, Barrea L, Sbardella E, Razzore P, Campione S, Faggiano A, Colao A; NIKE; Albertelli M, Altieri B, Bottiglieri F, De Cicco F, Di Molfetta S, Fanciulli G, Feola T, Ferone D, Ferraù F, Gallo M, Giannetta E, Grillo F, Grossrubatscher E, Guadagno E, Guarnotta V, Isidori AM, Lania A, Lenzi A, Calzo FL, Malandrino P, Messina E, Modica R, Muscogiuri G, Pes L, Pizza G, Pofi R, Puliani G, Rainone C, Rizza L, Rubino M, Ruggieri RM, Sesti F, Venneri MA, Zatelli MC. Vitale G, et al. Rev Endocr Metab Disord. 2021 Sep;22(3):511-525. doi: 10.1007/s11154-020-09589-y. Epub 2020 Sep 15. Rev Endocr Metab Disord. 2021. PMID: 32935263 Free PMC article. Review.
References
- Modlin I.M., et al. (2005). Current status of gastrointestinal carcinoids. Gastroenterology, 128, 1717–1751 - PubMed
- Valentino J., et al. (2011). Recent advances in the diagnosis and treatment of gastrointestinal carcinoids. Adv. Surg., 45, 285–300 - PubMed
- Modlin I.M., et al. (2003). A 5-decade analysis of 13,715 carcinoid tumors. Cancer, 97, 934–959 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources