Microbiome of the upper troposphere: species composition and prevalence, effects of tropical storms, and atmospheric implications - PubMed (original) (raw)
Microbiome of the upper troposphere: species composition and prevalence, effects of tropical storms, and atmospheric implications
Natasha DeLeon-Rodriguez et al. Proc Natl Acad Sci U S A. 2013.
Abstract
The composition and prevalence of microorganisms in the middle-to-upper troposphere (8-15 km altitude) and their role in aerosol-cloud-precipitation interactions represent important, unresolved questions for biological and atmospheric science. In particular, airborne microorganisms above the oceans remain essentially uncharacterized, as most work to date is restricted to samples taken near the Earth's surface. Here we report on the microbiome of low- and high-altitude air masses sampled onboard the National Aeronautics and Space Administration DC-8 platform during the 2010 Genesis and Rapid Intensification Processes campaign in the Caribbean Sea. The samples were collected in cloudy and cloud-free air masses before, during, and after two major tropical hurricanes, Earl and Karl. Quantitative PCR and microscopy revealed that viable bacterial cells represented on average around 20% of the total particles in the 0.25- to 1-μm diameter range and were at least an order of magnitude more abundant than fungal cells, suggesting that bacteria represent an important and underestimated fraction of micrometer-sized atmospheric aerosols. The samples from the two hurricanes were characterized by significantly different bacterial communities, revealing that hurricanes aerosolize a large amount of new cells. Nonetheless, 17 bacterial taxa, including taxa that are known to use C1-C4 carbon compounds present in the atmosphere, were found in all samples, indicating that these organisms possess traits that allow survival in the troposphere. The findings presented here suggest that the microbiome is a dynamic and underappreciated aspect of the upper troposphere with potentially important impacts on the hydrological cycle, clouds, and climate.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
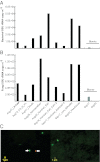
Quantification of bacterial and fungal cells in samples from high altitudes in the atmosphere. Concentration of bacterial (A) and fungal (B) cells based on qPCR analysis of SSU rRNA gene copies in the samples. Note that samples are ordered by the collection time on the x axis except for blank samples, which are shown at the rightmost part of the graphs in light gray. (C) Live/dead microscopy image of two samples from the California coast and transit flights. Green-stained cells represent cells with viable/intact membrane (e.g., cell indicated by left arrow), and red/yellow-stained cells represent cells with a damaged membrane (e.g., cell indicated by right arrow).
Fig. 2.
Composition of tropospheric bacterial communities. (A) Relative abundance (y axis) of taxa (see key) represented by the partial SSU rRNA gene sequences recovered in each sample (x axis). Black vertical lines next to the bars underline the core OTUs that are present in all samples. “*Others” refers to low-abundance OTUs; for a complete list of the OTUs grouped under “*Others,” refer to
SI Appendix, Table S3
. (B) Principal coordinates analysis based on the β-diversity values calculated by the weighted UniFrac distance metric of samples collected during the GRIP campaign and samples from previous studies (see key). Samples from this study are represented with open and closed squares and color-coded as follows: red, California/transit; blue, clouds; purple, low altitude; green, Hurricane Earl; orange, Hurricane Karl.
Fig. 3.
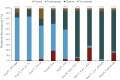
Habitat of origin of the SSU rRNA gene sequences recovered in the GRIP samples. Sequences were assigned to a habitat (see key) based on the source of isolation of their best match in the GreenGenes database. The graph represents the relative abundance of each habitat (vertical axis) for each sample (x axis). Numbers on the top denote the fraction of sequences that were assignable to a habitat for each sample. For a similar analysis that normalized for the differential representation of habitats among the reference GreenGenes sequences, see
SI Appendix, Fig. S6
.
Comment in
- Environmental microbiology: plant bacteria thrive in storm clouds.
Kåhrström CT. Kåhrström CT. Nat Rev Microbiol. 2013 Mar;11(3):146. doi: 10.1038/nrmicro2978. Nat Rev Microbiol. 2013. PMID: 23411853 No abstract available. - Inadequate methods and questionable conclusions in atmospheric life study.
Smith DJ, Griffin DW. Smith DJ, et al. Proc Natl Acad Sci U S A. 2013 Jun 4;110(23):E2084. doi: 10.1073/pnas.1302612110. Epub 2013 Apr 30. Proc Natl Acad Sci U S A. 2013. PMID: 23633574 Free PMC article. No abstract available. - Reply to Smith and Griffin: Methods, air flows, and conclusions are robust in the DeLeon-Rodriguez et al. study.
DeLeon-Rodriguez N, Lathem TL, Rodriguez-R LM, Barazesh JM, Anderson BE, Beyersdorf AJ, Ziemba LD, Bergin M, Nenes A, Konstantinidis KT. DeLeon-Rodriguez N, et al. Proc Natl Acad Sci U S A. 2013 Jun 4;110(23):E2085. doi: 10.1073/pnas.1304466110. Proc Natl Acad Sci U S A. 2013. PMID: 23882709 Free PMC article. No abstract available.
Similar articles
- Inadequate methods and questionable conclusions in atmospheric life study.
Smith DJ, Griffin DW. Smith DJ, et al. Proc Natl Acad Sci U S A. 2013 Jun 4;110(23):E2084. doi: 10.1073/pnas.1302612110. Epub 2013 Apr 30. Proc Natl Acad Sci U S A. 2013. PMID: 23633574 Free PMC article. No abstract available. - Seasonal variability in bacterial and fungal diversity of the near-surface atmosphere.
Bowers RM, Clements N, Emerson JB, Wiedinmyer C, Hannigan MP, Fierer N. Bowers RM, et al. Environ Sci Technol. 2013;47(21):12097-106. doi: 10.1021/es402970s. Epub 2013 Oct 22. Environ Sci Technol. 2013. PMID: 24083487 - Vertical stratification of the air microbiome in the lower troposphere.
Drautz-Moses DI, Luhung I, Gusareva ES, Kee C, Gaultier NE, Premkrishnan BNV, Lee CF, Leong ST, Park C, Yap ZH, Heinle CE, Lau KJX, Purbojati RW, Lim SBY, Lim YH, Kutmutia SK, Aung NW, Oliveira EL, Ng SG, Dacanay J, Ang PN, Spence S, Phung WJ, Wong A, Kennedy RJ, Kalsi N, Sasi SP, Chandrasekaran L, Uchida A, Junqueira ACM, Kim HL, Hankers R, Feuerle T, Corsmeier U, Schuster SC. Drautz-Moses DI, et al. Proc Natl Acad Sci U S A. 2022 Feb 15;119(7):e2117293119. doi: 10.1073/pnas.2117293119. Proc Natl Acad Sci U S A. 2022. PMID: 35131944 Free PMC article. - Tropospheric aerosols: size-differentiated chemistry and large-scale spatial distributions.
Hidy GM, Mohnen V, Blanchard CL. Hidy GM, et al. J Air Waste Manag Assoc. 2013 Apr;63(4):377-404. doi: 10.1080/10962247.2012.760499. J Air Waste Manag Assoc. 2013. PMID: 23687724 Review. - Monitoring of airborne biological particles in outdoor atmosphere. Part 1: Importance, variability and ratios.
Núñez A, Amo de Paz G, Rastrojo A, García AM, Alcamí A, Gutiérrez-Bustillo AM, Moreno DA. Núñez A, et al. Int Microbiol. 2016 Mar;19(1):1-13. doi: 10.2436/20.1501.01.258. Int Microbiol. 2016. PMID: 27762424 Review.
Cited by
- Inadequate methods and questionable conclusions in atmospheric life study.
Smith DJ, Griffin DW. Smith DJ, et al. Proc Natl Acad Sci U S A. 2013 Jun 4;110(23):E2084. doi: 10.1073/pnas.1302612110. Epub 2013 Apr 30. Proc Natl Acad Sci U S A. 2013. PMID: 23633574 Free PMC article. No abstract available. - Continental-Scale Gene Flow Prevents Allopatric Divergence of Pelagic Freshwater Bacteria.
Hoetzinger M, Pitt A, Huemer A, Hahn MW. Hoetzinger M, et al. Genome Biol Evol. 2021 Mar 1;13(3):evab019. doi: 10.1093/gbe/evab019. Genome Biol Evol. 2021. PMID: 33674852 Free PMC article. - Infection of phytoplankton by aerosolized marine viruses.
Sharoni S, Trainic M, Schatz D, Lehahn Y, Flores MJ, Bidle KD, Ben-Dor S, Rudich Y, Koren I, Vardi A. Sharoni S, et al. Proc Natl Acad Sci U S A. 2015 May 26;112(21):6643-7. doi: 10.1073/pnas.1423667112. Epub 2015 May 11. Proc Natl Acad Sci U S A. 2015. PMID: 25964340 Free PMC article. - Fog-Induced Alteration in Airborne Microbial Community: a Study over Central Indo-Gangetic Plain in India.
Saikh SR, Das SK. Saikh SR, et al. Appl Environ Microbiol. 2023 Jan 31;89(1):e0136722. doi: 10.1128/aem.01367-22. Epub 2023 Jan 9. Appl Environ Microbiol. 2023. PMID: 36622163 Free PMC article. - Airborne Microbial Communities at High-Altitude and Suburban Sites in Toyama, Japan Suggest a New Perspective for Bioprospecting.
Tanaka D, Sato K, Goto M, Fujiyoshi S, Maruyama F, Takato S, Shimada T, Sakatoku A, Aoki K, Nakamura S. Tanaka D, et al. Front Bioeng Biotechnol. 2019 Feb 5;7:12. doi: 10.3389/fbioe.2019.00012. eCollection 2019. Front Bioeng Biotechnol. 2019. PMID: 30805335 Free PMC article.
References
- Despres VR, et al. Primary biological aerosol particles in the atmosphere: A review. Tellus B Chem Phys Meterol. 2012;64 10.3402/tellusb.v64i0.15598.
- Möhler O, DeMott PJ, Vali G, Levin Z. Microbiology and atmospheric processes: The role of biological particles in cloud physics. Biogeosciences. 2007;4(6):1059–1071.
- Vali G, et al. Biogenic ice nuclei. Part II: Bacterial sources. J Atmos Sci. 1976;33(8):1565–1570.
- Bauer H, et al. Airborne bacteria as cloud condensation nuclei. J Geophys Res. 2003;108(D21):4658.
- Amato P. Clouds provide atmospheric oases for microbes. Microbe. 2012;7(3):119–123.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous