RNA-guided editing of bacterial genomes using CRISPR-Cas systems - PubMed (original) (raw)
RNA-guided editing of bacterial genomes using CRISPR-Cas systems
Wenyan Jiang et al. Nat Biotechnol. 2013 Mar.
Abstract
Here we use the clustered, regularly interspaced, short palindromic repeats (CRISPR)-associated Cas9 endonuclease complexed with dual-RNAs to introduce precise mutations in the genomes of Streptococcus pneumoniae and Escherichia coli. The approach relies on dual-RNA:Cas9-directed cleavage at the targeted genomic site to kill unmutated cells and circumvents the need for selectable markers or counter-selection systems. We reprogram dual-RNA:Cas9 specificity by changing the sequence of short CRISPR RNA (crRNA) to make single- and multinucleotide changes carried on editing templates. Simultaneous use of two crRNAs enables multiplex mutagenesis. In S. pneumoniae, nearly 100% of cells that were recovered using our approach contained the desired mutation, and in E. coli, 65% that were recovered contained the mutation, when the approach was used in combination with recombineering. We exhaustively analyze dual-RNA:Cas9 target requirements to define the range of targetable sequences and show strategies for editing sites that do not meet these requirements, suggesting the versatility of this technique for bacterial genome engineering.
Conflict of interest statement
The authors have no conflicting financial interests.
Figures
Figure 1
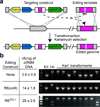
Cas9 nuclease activity against endogenous targets can be exploited for genome editing. (a) Concept of CRISPR directed genome editing. The CRISPR targeting construct directs interference against a chromosomal locus and is co-transformed with an editing template that recombines with the target to abolish the interference. Kanamycin-resistant transformants that survive CRISPR attack contain modifications introduced by the editing template. (b) Transformation of crR6M DNA in R68232.5 cells with either: no editing template, the R6 srtA or the R6370.1 editing templates. Transformation efficiency is calculated as colony forming units (cfu) per µg of crR6M DNA; the mean values with standard deviations from at least three independent experiments are shown. PCR analysis is performed on 8 clones in each transformation. “Un.” indicates the unedited srtA locus of strain R68232.5; “Ed.” shows the editing template. R68232.5 and R6370.1 targets are distinguished by restriction with EaeI.
Figure 2
Analysis of PAM and seed sequences that eliminate CRISPR interference. (a) PCR products with randomized PAM sequences or randomized seed sequences (in green) were transformed in crR6 cells. These cells express Cas9 loaded with a crRNA that targets a chromosomal region of R68232.5 cells (highlighted in pink) that is absent from the R6 genome. More than 2×105 chloramphenicol-resistant transformants, carrying inactive PAM or seed sequences, were combined for amplification and deep sequencing of the target region. (b) Relative proportion of number of reads after transformation of the random PAM constructs in crR6 cells (compared to number of reads in R6 transformants). The relative abundance for each 3-nucleotide PAM sequence is shown. Severely underrepresented sequences (NGG) are shown in red; partially underrepresented one in orange (NAG) (c) Relative proportion of number of reads after transformation of the random seed sequence constructs in crR6 cells (compared to number of reads in R6 transformants). The relative abundance of each nucleotide for each position of the first 20 nucleotides of the protospacer sequence is shown. High abundance indicates lack of CRISPR targeting, i.e. a CRISPR inactivating mutation. The grey line shows the level of the WT sequence. The dotted line represents the level above which a mutation significantly disrupts interference (Supplementary text).
Figure 3
Introduction of single and multiple mutations using CRISPR-mediated genome editing. (a) Nucleotide and amino acid sequences of the wild-type and edited (green nucleotides; underlined amino acid residues) bgaA. The protospacer, PAM and restriction sites are shown. (b) Transformation efficiency of cells transformed with targeting constructs in the presence of an editing template or control. (c) PCR analysis for 8 transformants of each editing experiment followed by digestion with BtgZI (R>A) and TseI (NE>AA). Deletion of bgaA is revealed as a smaller PCR product. (d) Miller assay to measure the β-galactosidase activity of WT and edited strains. (e) For a single-step double deletion the targeting construct contains two spacers (in this case matching srtA and bgaA) and is co-transformed with two different editing templates (f) PCR analysis for 8 transformants to detect deletions in srtA and bgaA loci. 6/8 transformants contained deletions of both genes.
Figure 4
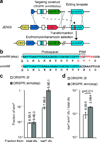
Mechanisms underlying CRISPR directed editing. (a) A stop codon was introduced in the erythromycin resistance gene ermAM to generate strain JEN53. The wild-type sequence can be restored through CRISPR-directed editing by targeting the stop codon with the CRISPR::ermAM(stop) construct, and using the ermAM wild-type sequence as an editing template. (b) Mutant and wild-type ermAM sequences. (c) Fraction of erythromicyn-resistant (ermR) cfu calculated from total or kanamycin-resistant (kanR) cfu, in the presence or absence of CRISPR targeting. (d) Fraction of total cells that acquire both the CRISPR construct and the editing template, in the presence or absence of CRISPR targeting. Co-transformation of the CRISPR targeting construct produced more transformants: t-test, t=0.011. In all cases the values show the mean±s.d. for three independent experiments.
Figure 5
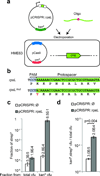
CRISPR-directed editing in E. coli. (a) A pCRISPR plasmid targeting the gene to edit can be transformed in the HME63 recombineering strain containing pCas9, together with an oligonucleotide specifying the mutation. (b) A K42T mutation conferring streptomycin resistance was introduced in the rpsL gene (c) Fraction of streptomicyn-resistant (strepR) cfu calculated from total or kanamycin-resistant (kanR) cfu, in the presence or absence of CRISPR targeting. (d) Fraction of total cells that acquire both the pCRISPR plasmid and the editing oligonucleotide, in the presence or absence of CRISPR targeting. Co-transformation of the pCRISPR targeting plasmid produced more transformants: t-test, t=0.004. In all cases the values show the mean±s.d. for three independent experiments.
Similar articles
- Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease.
Cho SW, Kim S, Kim JM, Kim JS. Cho SW, et al. Nat Biotechnol. 2013 Mar;31(3):230-2. doi: 10.1038/nbt.2507. Epub 2013 Jan 29. Nat Biotechnol. 2013. PMID: 23360966 - Efficient genome editing in zebrafish using a CRISPR-Cas system.
Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Hwang WY, et al. Nat Biotechnol. 2013 Mar;31(3):227-9. doi: 10.1038/nbt.2501. Epub 2013 Jan 29. Nat Biotechnol. 2013. PMID: 23360964 Free PMC article. - Selection and Validation of Spacer Sequences for CRISPR-Cas9 Genome Editing and Transcription Regulation in Bacteria.
Grenier F, Lucier JF, Rodrigue S. Grenier F, et al. Methods Mol Biol. 2015;1334:233-44. doi: 10.1007/978-1-4939-2877-4_15. Methods Mol Biol. 2015. PMID: 26404154 - Programmable DNA cleavage in vitro by Cas9.
Karvelis T, Gasiunas G, Siksnys V. Karvelis T, et al. Biochem Soc Trans. 2013 Dec;41(6):1401-6. doi: 10.1042/BST20130164. Biochem Soc Trans. 2013. PMID: 24256227 Review. - Cas9 as a versatile tool for engineering biology.
Mali P, Esvelt KM, Church GM. Mali P, et al. Nat Methods. 2013 Oct;10(10):957-63. doi: 10.1038/nmeth.2649. Nat Methods. 2013. PMID: 24076990 Free PMC article. Review.
Cited by
- Target specificity of the CRISPR-Cas9 system.
Wu X, Kriz AJ, Sharp PA. Wu X, et al. Quant Biol. 2014 Jun;2(2):59-70. doi: 10.1007/s40484-014-0030-x. Quant Biol. 2014. PMID: 25722925 Free PMC article. - Current updates of CRISPR/Cas9-mediated genome editing and targeting within tumor cells: an innovative strategy of cancer management.
Allemailem KS, Alsahli MA, Almatroudi A, Alrumaihi F, Alkhaleefah FK, Rahmani AH, Khan AA. Allemailem KS, et al. Cancer Commun (Lond). 2022 Dec;42(12):1257-1287. doi: 10.1002/cac2.12366. Epub 2022 Oct 9. Cancer Commun (Lond). 2022. PMID: 36209487 Free PMC article. Review. - Mechanistic toxicology in light of genetic compensation.
Elizalde MJ, Gorelick DA. Elizalde MJ, et al. Toxicol Sci. 2023 Nov 6;197(2):115-20. doi: 10.1093/toxsci/kfad113. Online ahead of print. Toxicol Sci. 2023. PMID: 37941503 Free PMC article. - On the ever-growing functional versatility of the CRISPR-Cas13 system.
Montagud-Martínez R, Márquez-Costa R, Heras-Hernández M, Dolcemascolo R, Rodrigo G. Montagud-Martínez R, et al. Microb Biotechnol. 2024 Feb;17(2):e14418. doi: 10.1111/1751-7915.14418. Microb Biotechnol. 2024. PMID: 38381083 Free PMC article. Review. - Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands.
Vercoe RB, Chang JT, Dy RL, Taylor C, Gristwood T, Clulow JS, Richter C, Przybilski R, Pitman AR, Fineran PC. Vercoe RB, et al. PLoS Genet. 2013 Apr;9(4):e1003454. doi: 10.1371/journal.pgen.1003454. Epub 2013 Apr 18. PLoS Genet. 2013. PMID: 23637624 Free PMC article.
References
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. - PubMed
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. - PubMed
- Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. - PubMed
- Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1DP2AI104556-01/AI/NIAID NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- DP1 MH100706/MH/NIMH NIH HHS/United States
- DP2 AI104556/AI/NIAID NIH HHS/United States
- R01/PHS HHS/United States
- DP1MH100706/DP/NCCDPHP CDC HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials