The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse - PubMed (original) (raw)
The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse
Phillip Grote et al. Dev Cell. 2013.
Abstract
The histone-modifying complexes PRC2 and TrxG/MLL play pivotal roles in determining the activation state of genes controlling pluripotency, lineage commitment, and cell differentiation. Long noncoding RNAs (lncRNAs) can bind to either complex, and some have been shown to act as modulators of PRC2 or TrxG/MLL activity. Here we show that the lateral mesoderm-specific lncRNA Fendrr is essential for proper heart and body wall development in the mouse. Embryos lacking Fendrr displayed upregulation of several transcription factors controlling lateral plate or cardiac mesoderm differentiation, accompanied by a drastic reduction in PRC2 occupancy along with decreased H3K27 trimethylation and/or an increase in H3K4 trimethylation at their promoters. Fendrr binds to both the PRC2 and TrxG/MLL complexes, suggesting that it acts as modulator of chromatin signatures that define gene activity. Thus, we identified an lncRNA that plays an essential role in the regulatory networks controlling the fate of lateral mesoderm derivatives.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
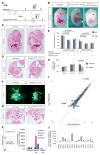
Figure 1. Fendrr is transiently expressed in nascent lateral plate mesoderm of developing mouse embryos
(A) Schematic of the genomic region of Fendrr and Foxf1. (B) Whole-mount in situ hybridization of mouse embryos at E8.25 and E9.5 revealed that Fendrr is transiently expressed and restricted to nascent lateral plate mesoderm (LPM), while Foxf1 transcripts persist in differentiating LPM along the body axis.
Figure 2. Heart and body wall development are impaired in mutants lacking Fendrr transcripts
(A) Schematic showing the wild type (upper) and targeted (lower) Fendrr-Foxf1 genomic regions; the first exon of Fendrr was replaced with a 3xpA stop cassette by gene targeting in order to prevent transcription of the gene. Arrows indicate the position of primers used for real-time qPCR analysis. (B) E13.75 embryos derived by tetraploid complementation from wild type or homozygous Fendrr mutant ES cells including the neighbouring selection cassettes (Fendrr3xpA[N)/3xpA(H]). In mutant embryos the ventral body wall is perforated by umbilical vessels and parts of the liver; the presence of selection cassettes had no visible effect on the phenotype (see also Figure S3A and S3B). The phenotype was rescued by expression of Fendrr from a modified BAC transgene (Tg(RP23-455G4)Bgh). Note that the tail and limbs have been removed to better observe the ventral malformations. Scale bar: 1 mm. (C) Transverse histological sections of E12.5 wild type and mutant embryos at the mid-trunk level. The body wall of E12.5 mutant embryos is significantly thinner than that of wild type embryos (arrowheads and inlet), leading to perforation by the liver and umbilical vessels at later stages. (D) Transverse histological sections of E12.5 wild type and mutant embryos at the chest level. The thickness of the ventricular wall in the mutant heart is significantly reduced as compared to wild type (inlet). Abbreviations: rv, right ventricle; is, interventricular septum; cu, atrio-ventricular endocardial cushion; la, left atrium; lv, left ventricle. Scale bar: 200 μm. (E) Tissue thickness was measured from Eosin-stained transverse sections of E12.5 wild type and homozygous mutant embryos. Measurements were taken on either side using Zeiss AxioVision software, and values from both sides were combined for paired t-test analysis. Mean +/− s.d. are shown (n=3). (F) Percentage of mitotic cells (H3S10P) in heart ventricles determined on two distinct sections each of three different embryos. Paired t-test analysis and mean +/− s.d. are shown (n=3).
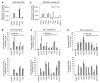
Figure 3. Loss of Fendrr affects epigenetic promoter modification and expression of transcriptional regulators in E8.5 embryonic hearts and caudal ends
(A, C) Normalized expression levels of transcriptional regulators in heart fields (A) and caudal ends (C) of Fendrr mutant embryos compared to wild type controls at the developmental stages listed. Several control genes are up-regulated in the mutants. n.d. = not detectable; * p≤0.02 in E8.5, ** p≤0.05 in E9.5. (B, D) Quantitative comparison of the levels of activating H3K4me3 and repressive H3K27me3 methylation marks in the promoters of control genes in E8.5 heart fields (B) and caudal ends (D) from Fendrr mutant and wild type embryos. H3K4 tri-methylation is increased at promoters of genes showing increased expression in the mutants (* p≤0.03). In addition, H3K27 tri-methylation is decreased at the promoters of control genes for LPM differentiation (Foxf1, Pitx2, Irx3) (* p≤0.05). (E) Quantitative comparison of the levels of WDR5 or EZH2 bound to the promoters of control factors in Fendrr mutant or wild type caudal ends. EZH2 occupancy at the Foxf1, Irx3 and Pitx2 promoters is dramatically reduced in the mutant, while other promoters are not affected. WDR5 occupancy is not altered for any of the promoters tested. All measurements were performed by quantitative real-time qPCR. Means +/− s.d. are shown (n=3 for expression and n=2 for ChIP analysis) (* p≤0.05).
Figure 4. Fendrr binds to the PRC2 and TrxG/MLL complexes and to target promoters
(A, B) RNA co-immunoprecipitation (RIP) from forebrain (upper panels) and caudal end (lower panels) lysates from wild type embryos using antibodies directed against the PRC2 components EZH2 and SUZ12 (A) or the TrxG/MLL component WDR5 (B); normal rabbit IgG was used as a control. Fold enrichment has been normalized to non-enriched input sample and U1 rRNA. Fendrr transcripts co-precipitated with EZH2, SUZ12, and WDR5 from caudal end tissue only, while the control lncRNA Hotair co-precipitated only with EZH2 and SUZ12 and HOTTIP lncRNA co-precipitated only with WDR5 from the caudal end tissue. Foxf1 and Hmbs RNA co-precipitation was used as a negative control. Mean +/− s.d. are shown (n=3). (C) Binding potential between Fendrr and genomic regions. The red curve shows the average probability of single stranded RNA as computed by sfold with a length parameter of 200 and W=1 (Ding et al. 2004). The heat map represents the base-pairing energy for an RNA/RNA duplex model for 40bp regions along the Fendrr transcript and 2,000 bp around the TSS of Foxf1 (top) and Pitx2 (bottom). The duplex energy is computed for each such region, staggered by 20bp. The optimal base pairing occurs in a region of Fendrr that also strongly favors single-stranded RNA, suggesting an open conformation that would enable the binding to the promoter. Probabilities are then averaged for a sliding window of 40bp to give the average RNA accessibility of the region that is binding. (D) Representation of the predicted interaction of the Fendrr RNA region and the promoter DNA region exhibiting the lowest free energy of approximately −70 kcal/mol (see yellow spot in C). (E) In vitro RNA/dsDNA binding assay utilizing biotin tagged RNA oligos as bait. Bars represent normalized enrichment of indicated 2,000 bp promoter fragment over background using a control RNA oligonucleotide (n=3, Mean +/− s.d. is shown).
Similar articles
- The long non-coding RNA Fendrr links epigenetic control mechanisms to gene regulatory networks in mammalian embryogenesis.
Grote P, Herrmann BG. Grote P, et al. RNA Biol. 2013 Oct;10(10):1579-85. doi: 10.4161/rna.26165. Epub 2013 Aug 22. RNA Biol. 2013. PMID: 24036695 Free PMC article. - Association of the Long Non-coding RNA Steroid Receptor RNA Activator (SRA) with TrxG and PRC2 Complexes.
Wongtrakoongate P, Riddick G, Fucharoen S, Felsenfeld G. Wongtrakoongate P, et al. PLoS Genet. 2015 Oct 23;11(10):e1005615. doi: 10.1371/journal.pgen.1005615. eCollection 2015 Oct. PLoS Genet. 2015. PMID: 26496121 Free PMC article. - Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol.
Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Bhan A, et al. J Mol Biol. 2013 Oct 9;425(19):3707-22. doi: 10.1016/j.jmb.2013.01.022. Epub 2013 Jan 31. J Mol Biol. 2013. PMID: 23375982 Free PMC article. - The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2.
Davidovich C, Cech TR. Davidovich C, et al. RNA. 2015 Dec;21(12):2007-22. doi: 10.1261/rna.053918.115. RNA. 2015. PMID: 26574518 Free PMC article. Review. - Long Non-Coding RNA FENDRR: Gene Structure, Expression, and Biological Relevance.
Szafranski P, Stankiewicz P. Szafranski P, et al. Genes (Basel). 2021 Jan 27;12(2):177. doi: 10.3390/genes12020177. Genes (Basel). 2021. PMID: 33513839 Free PMC article. Review.
Cited by
- Unveiling dysregulated lncRNAs and networks in non-syndromic cleft lip with or without cleft palate pathogenesis.
Wu C, Liu H, Zhan Z, Zhang X, Zhang M, You J, Ma J. Wu C, et al. Sci Rep. 2024 Jan 10;14(1):1047. doi: 10.1038/s41598-024-51747-8. Sci Rep. 2024. PMID: 38200098 Free PMC article. - Epigenetics of the failing heart.
Marín-García J, Akhmedov AT. Marín-García J, et al. Heart Fail Rev. 2015 Jul;20(4):435-59. doi: 10.1007/s10741-015-9483-x. Heart Fail Rev. 2015. PMID: 25847519 Review. - The centrosomal protein 83 (CEP83) regulates human pluripotent stem cell differentiation toward the kidney lineage.
Mansour F, Hinze C, Telugu NS, Kresoja J, Shaheed IB, Mosimann C, Diecke S, Schmidt-Ott KM. Mansour F, et al. Elife. 2022 Oct 12;11:e80165. doi: 10.7554/eLife.80165. Elife. 2022. PMID: 36222666 Free PMC article. - Long noncoding RNA LYPLAL1-AS1 regulates adipogenic differentiation of human mesenchymal stem cells by targeting desmoplakin and inhibiting the Wnt/β-catenin pathway.
Yang Y, Fan J, Xu H, Fan L, Deng L, Li J, Li D, Li H, Zhang F, Zhao RC. Yang Y, et al. Cell Death Discov. 2021 May 15;7(1):105. doi: 10.1038/s41420-021-00500-5. Cell Death Discov. 2021. PMID: 33993187 Free PMC article. - Androgen Receptor-Related Non-coding RNAs in Prostate Cancer.
Yang Y, Liu KY, Liu Q, Cao Q. Yang Y, et al. Front Cell Dev Biol. 2021 Apr 1;9:660853. doi: 10.3389/fcell.2021.660853. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33869227 Free PMC article. Review.
References
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–8. - PubMed
- Besch R, Giovannangeli C, Schuh T, Kammerbauer C, Degitz K. Characterization and quantification of triple helix formation in chromosomal DNA. Journal of molecular biology. 2004;341:979–89. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases