Msx1 homeodomain protein represses the αGSU and GnRH receptor genes during gonadotrope development - PubMed (original) (raw)
Msx1 homeodomain protein represses the αGSU and GnRH receptor genes during gonadotrope development
Huimin Xie et al. Mol Endocrinol. 2013 Mar.
Abstract
Multiple homeodomain transcription factors are crucial for pituitary organogenesis and cellular differentiation. A homeodomain repressor, Msx1, is expressed from the ventral aspect of the developing anterior pituitary and implicated in gonadotrope differentiation. Here, we find that Msx1 represses transcription of lineage-specific pituitary genes such as the common α-glycoprotein subunit (αGSU) and GnRH receptor (GnRHR) promoters in the mouse gonadotrope-derived cell lines, αT3-1 and LβT2. Repression of the mouse GnRHR promoter by Msx1 is mediated through a consensus-binding motif in the downstream activin regulatory element (DARE). Truncation and mutation analyses of the human αGSU promoter map Msx1 repression to a site at -114, located at the junctional regulatory element (JRE). Dlx activators are closely related to the Msx repressors, acting through the same elements, and Dlx3 and Dlx2 act as transcriptional activators for GnRHR and αGSU, respectively. Small interfering RNA knockdown of Msx1 in αT3-1 cells increases endogenous αGSU and GnRHR mRNA expression. Msx1 gene expression reaches its maximal expression at the rostral edge at e13.5. The subsequent decline in Msx1 expression specifically coincides with the onset of expression of both αGSU and GnRHR. The expression levels of both αGSU and GnRHR in Msx1-null mice at e18.5 are higher compared with wild type, further confirming a role for Msx1 in the repression of αGSU and GnRHR. In summary, Msx1 functions as a negative regulator early in pituitary development by repressing the gonadotrope-specific αGSU and GnRHR genes, but a temporal decline in Msx1 expression alleviates this repression allowing induction of GnRHR and αGSU, thus serving to time the onset of gonadotrope-specific gene program.
Figures
Figure 1.
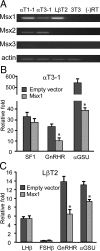
Msx1 represses transcription of GnRHR and αGSU promoters in gonadotropes. (A) Msx mRNA expression in the gonadotrope-derived cell lines. Total RNA was harvested from the mouse gonadotrope-derived cell lines: progenitor αT1-1, immature αT3-1, mature LβT2, and NIH 3T3 cells as control, then reverse transcribed to generate cDNA. β-Actin was used as loading control. (B) The wild-type rat SF1, human αGSU, and mouse GnRHR-luciferase reporters were cotransfected with Msx1 expression vector or an equal mass of empty expression vector into αT3-1 cells. (C) The wild-type FSHβ, LHβ, human αGSU, mouse GnRHR, and pGL3 luciferase reporters were cotransfected with Msx1 expression vector or an equal mass of empty expression vector in LβT2 cells. Abbreviation: RT, reverse transcriptase. An asterisk indicates significant differences (P < .05) from empty vector control.
Figure 2.
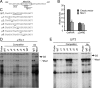
Msx1 represses GnRHR promoter through DARE element. (A) Schematic of DARE element on mouse GnRHR promoter. Box indicates the putative Msx1/Dlx-binding site. The Lhx3, Oct-1, and NF-Y sites on the DARE element are underlined. (B) Msx1 represses GnRHR transcription through DARE element. The wild-type GnRHR and DARE mutant (μDARE) reporters were transiently transfected into αT3-1 cells. The pGL3 empty luciferase reporter was used as control. Cells were cotransfected with Msx1 expression vector or an equal mass of empty expression vector. (C) The oligonucleotides used as the DARE probe and its mutants (μ1–μ6) are listed, and the Msx1-binding element and the 2-bp transversions are underlined. The binding of Msx1 on the GnRHR promoter maps to the DARE element using αT3-1 (D) and LβT2 (E) gonadotrope nuclear extracts. Specific protein/DNA complexes were identified by competition with 1000-fold excess unlabeled oligonucleotides, including homologous oligonucleotides or ones containing a defined Msx1-binding site from the rat GnRH promoter (positive control) or the 2-bp transversions. Confirmation of the presence of Msx1 in specific complexes was determined by inclusion of an antibody directed against Msx1 or mouse IgG as a control. The hollow arrow marks the Msx1 complex, and the arrow marks the supershift of the Msx1 complex after binding with Msx1 antibody. SS represents supershift. Abbreviations: WT, wild type; Ab, antibody.
Figure 3.
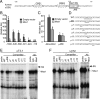
Msx1 represses the αGSU promoter at the −114 JRE within −224 bp of αGSU mRNA start site. (A) Gonadotrope regulatory elements in the proximal human αGSU promoter. Thick boxes indicate the putative Msx1/Dlx-binding elements, the −114 site. The JRE element is underlined. CRE1 and CRE2 are marked with dashed ovals. (B) The full-length human αGSU promoter and its truncated reporters map Msx1 repression inside of −224 bp of the start site on the αGSU promoter. The wild-type −1800 αGSU-luc, and its truncations: −845 αGSU-luc, −668 αGSU-luc, −391 αGSU-luc, −224 αGSU-luc, and −116 αGSU-luc, were transiently transfected into αT3-1 cells with the Msx1 expression vector or an equal mass of empty expression vector. (C) Identification of Msx1-binding sites on the human αGSU promoter. Wild-type −224 αGSU reporter (WT) and mutants (μJRE) were transiently transfected into αT3-1 cells, pGL3 empty luciferase reporters were used as control. Cells were cotransfected with Msx1 or an equal mass of empty expression vector. Groups labeled with different letters are significantly different from each other (P < .05). (D) The oligonucleotides used for the αGSU JRE probe and its mutants (μ7–μ12) are listed, and the Msx1-binding element and 2-bp transversions are underlined. Msx1 binding on the αGSU promoter maps to the −114 site in αT3-1 (E) and LβT2 (F) gonadotropes. The hollow arrow marks the Msx1 complex, and the arrow marks the supershift of the Msx1 complex after binding with Msx1 antibody. SS represents supershift complex. Abbreviation: Ab, antibody.
Figure 4.
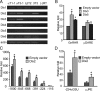
Dlx proteins are activators of both αGSU and GnRHR transcription. (A) Dlx mRNA expression in the gonadotrope-derived cell lines. Total RNA was harvested from gonadotrope-derived cell lines: progenitor αT1-1, immature αT3-1, mature LβT2, and NIH 3T3 cells as control, then reverse transcribed to generate cDNA. β-Actin was used as loading control. Resulting PCR reactions were run on a 1.5% agarose gel. (B) Dlx3 and Dlx5 activate GnRHR transcription through the DARE element. Wild-type GnRHR and DARE mutant (μDARE) reporters were transiently transfected into αT3-1 cells, pGL3 luciferase reporters were used as control. Cells were cotransfected with Dlx expression vector or an equal mass of empty expression vector. (C) Dlx2 activates αGSU transcription, and this effect maps to inside of −224 bp on the αGSU promoter. The wild-type −1800 αGSU-luciferase and its truncations were transiently transfected into the αT3-1 cell line with Dlx2 expression vector or an equal mass of empty expression vector. (D) Identification of Dlx2-binding sites on αGSU promoter. Wild-type −224 αGSU and mutant (μJRE) reporters were transiently transfected into αT3-1 cells, pGL3 luciferase reporters were used as control. Cells were cotransfected with Dlx2 expression vector or an equal mass of empty expression vector. Abbreviation: RT, reverse transcriptase. An asterisk indicates significant differences (P < .05) from empty vector control.
Figure 5.
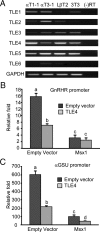
Msx1 recruits TLE4 as a corepressor to repress αGSU and GnRHR transcription. (A) TLE expression in the gonadotrope-derived cell lines. Total RNAs were harvested from progenitor αT1-1, immature αT3-1, mature LβT2, and fibroblast NIH 3T3 cells, then reverse transcribed to generate cDNA. GAPDH was used as loading control. (B and C) TLE4 is involved in Msx1-mediated GnRHR (B) and αGSU (C) repression. pGL3 empty luciferase reporters were used as control. Cells were cotransfected with Msx1 expression vector or an equal mass of empty expression vector, with or without TLE4 expression vector. Groups labeled with different letters are significantly different from each other (P < .05). Abbreviation: RT, reverse transcriptase.
Figure 6.
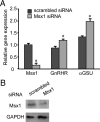
Knockdown of endogenous Msx1 in αT3-1 cells increases endogenous mouse αGSU and GnRHR mRNAs. (A) αT3-1 cells were transfected with SMARTpools of siRNA for Msx1 or a scrambled control. Cells were transfected for 72 hours, at which point total RNA were harvested and reverse transcribed. The resulting cDNA was used in qPCR reactions with primers specific for Msx1, αGSU, GnRHR, and GAPDH. All values represent the SQ means from 4 independent experiments with all means adjusted to corresponding GAPDH values within experiment. All values are normalized to scrambled control. (B) Whole-cell lysates were used for protein expression by Western blotting. An asterisk indicates significant differences (P < .05) from empty vector control.
Figure 7.
Msx1/β-gal expression peaks during mouse anterior pituitary development on e13.5 and begins to decline by e15.5 concurrent with increased αGSU expression. Embryos produced by mating heterozygote Msx1/βgal males and females were fixed, embedded, sectioned, and subjected to immunohistochemical. (A–D) Schematic diagrams of pituitary development at different stages. Rathke's pouch (RP) is formed and makes direct contact with the portion of the ventral diencephalon (VD) comprising the infundibulum (INF). Five different pituitary cell types are hypothesized to proliferate ventrally from the epithelial cells of RP to populate the anterior lobe of the pituitary gland. Wild-type embryos (E–H) and _Msx1_-null/β-gal insertion (I–L) illustrate sections of e11.5, e13.5, e15.5, and e17.5 developing mouse anterior pituitaries with methyl green as a counter stain, whereas the purple/red peroxidase staining represents β-gal in the place of normal Msx1 expression. (M and N) Brown DAB (3,3'-diaminobenzidine) staining representing αGSU expression in wild-type embryos at e13.5 and e15.5, respectively. The arrow indicates the rostral tip of the developing anterior pituitary.
Figure 8.
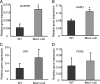
GnRHR and αGSU mRNA expressions in developing pituitary are highly induced in _Msx1_-null mice. Embryo of both wild-type (WT) and null mice at e18.5 were harvested by timed mating of heterozygote Msx1/β-gal males and females. Total RNAs were isolated from pituitaries and then reverse transcribed to generate cDNA. qPCRs were performed with specific primers for (A) GnRHR, (B) αGSU, (C) LHβ, and (D) FSHβ. Values represent the SQ means from 3 (A and B) or 5 (C and D) groups of embryonic pituitaries, and each group contains 5 pituitaries, with all means adjusted to corresponding GAPDH values within experiment. An asterisk represents values (P < .05) different from wild type (+/+).
Similar articles
- Glucocorticoids induce human glycoprotein hormone alpha-subunit gene expression in the gonadotrope.
Sasson R, Luu SH, Thackray VG, Mellon PL. Sasson R, et al. Endocrinology. 2008 Jul;149(7):3643-55. doi: 10.1210/en.2007-1100. Epub 2008 Apr 10. Endocrinology. 2008. PMID: 18403486 Free PMC article. - Msx1 homeodomain transcription factor and TATA-binding protein interact to repress the expression of the glycoprotein hormone α subunit gene.
Park KS, Kim KK, Kim KE. Park KS, et al. Biochem Biophys Res Commun. 2015 Dec 4-11;468(1-2):326-30. doi: 10.1016/j.bbrc.2015.10.102. Epub 2015 Oct 23. Biochem Biophys Res Commun. 2015. PMID: 26505791 - GATA2-induced silencing and LIM-homeodomain protein-induced activation are mediated by a bi-functional response element in the rat GnRH receptor gene.
Schang AL, Granger A, Quérat B, Bleux C, Cohen-Tannoudji J, Laverrière JN. Schang AL, et al. Mol Endocrinol. 2013 Jan;27(1):74-91. doi: 10.1210/me.2012-1182. Epub 2012 Dec 4. Mol Endocrinol. 2013. PMID: 23211524 Free PMC article. - [An ambiguous role of steroidogenic factor 1 in the rat GnRH receptor gene expression. Lessons from transgenic mice].
Laverrière JN, Granger A, Pincas H, Ngô-Muller V, Bleux C, Tixier-Vidal A, Magre S, Guigon C, Daegelen D, Counis R. Laverrière JN, et al. J Soc Biol. 2004;198(1):73-9. J Soc Biol. 2004. PMID: 15146959 Review. French. - Regulation of expression of mammalian gonadotrophin-releasing hormone receptor genes.
Hapgood JP, Sadie H, van Biljon W, Ronacher K. Hapgood JP, et al. J Neuroendocrinol. 2005 Oct;17(10):619-38. doi: 10.1111/j.1365-2826.2005.01353.x. J Neuroendocrinol. 2005. PMID: 16159375 Review.
Cited by
- Homeodomain Proteins SIX3 and SIX6 Regulate Gonadotrope-specific Genes During Pituitary Development.
Xie H, Hoffmann HM, Meadows JD, Mayo SL, Trang C, Leming SS, Maruggi C, Davis SW, Larder R, Mellon PL. Xie H, et al. Mol Endocrinol. 2015 Jun;29(6):842-55. doi: 10.1210/me.2014-1279. Epub 2015 Apr 27. Mol Endocrinol. 2015. PMID: 25915183 Free PMC article. - The _Cis_-Regulatory Code for Kelch-like 21/30 Specific Expression in Ciona robusta Sensory Organs.
Coppola U, Kamal AK, Stolfi A, Ristoratore F. Coppola U, et al. Front Cell Dev Biol. 2020 Sep 11;8:569601. doi: 10.3389/fcell.2020.569601. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33043001 Free PMC article. - WDR11-mediated Hedgehog signalling defects underlie a new ciliopathy related to Kallmann syndrome.
Kim YJ, Osborn DP, Lee JY, Araki M, Araki K, Mohun T, Känsäkoski J, Brandstack N, Kim HT, Miralles F, Kim CH, Brown NA, Kim HG, Martinez-Barbera JP, Ataliotis P, Raivio T, Layman LC, Kim SH. Kim YJ, et al. EMBO Rep. 2018 Feb;19(2):269-289. doi: 10.15252/embr.201744632. Epub 2017 Dec 20. EMBO Rep. 2018. PMID: 29263200 Free PMC article. - Fshb-iCre mice are efficient and specific Cre deleters for the gonadotrope lineage.
Wang H, Hastings R, Miller WL, Kumar TR. Wang H, et al. Mol Cell Endocrinol. 2016 Jan 5;419:124-38. doi: 10.1016/j.mce.2015.10.006. Epub 2015 Oct 22. Mol Cell Endocrinol. 2016. PMID: 26472536 Free PMC article. - A human ACTH-secreting corticotroph tumoroid model: Novel Human ACTH-Secreting Tumor Cell in vitro Model.
Zhang D, Hugo W, Redublo P, Miao H, Bergsneider M, Wang MB, Kim W, Yong WH, Heaney AP. Zhang D, et al. EBioMedicine. 2021 Apr;66:103294. doi: 10.1016/j.ebiom.2021.103294. Epub 2021 Mar 25. EBioMedicine. 2021. PMID: 33773184 Free PMC article.
References
- Simmons DM, Voss JW, Ingraham HA, et al. Pituitary cell phenotypes involve cell-specific Pit-1 mRNA translation and synergistic interactions with other classes of transcription factors. Genes Dev. 1990;4:695–711 - PubMed
- Scully KM, Rosenfeld MG. Pituitary development: regulatory codes in mammalian organogenesis. Science. 2002;295:2231–2235 - PubMed
- Windle JJ, Weiner RI, Mellon PL. Cell lines of the pituitary gonadotrope lineage derived by targeted oncogenesis in transgenic mice. Mol Endocrinol. 1990;4:597–603 - PubMed
- Alarid ET, Windle JJ, Whyte DB, Mellon PL. Immortalization of pituitary cells at discrete stages of development by directed oncogenesis in transgenic mice. Development. 1996;122:3319–3329 - PubMed
- Lew D, Brady H, Klausing K, et al. GHF-1 promoter-targeted immortalization of a somatotropic progenitor cell results in dwarfism in transgenic mice. Genes Dev. 1993;7:683–693 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HD072754/HD/NICHD NIH HHS/United States
- P42 ES101337/ES/NIEHS NIH HHS/United States
- T32 HD007203/HD/NICHD NIH HHS/United States
- R01 HD020377/HD/NICHD NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- P50 HD028934/HD/NICHD NIH HHS/United States
- R37 HD020377/HD/NICHD NIH HHS/United States
- R01 DK044838/DK/NIDDK NIH HHS/United States
- T32 GM008666/GM/NIGMS NIH HHS/United States
- P50 HD012303/HD/NICHD NIH HHS/United States
- U54 HD012303/HD/NICHD NIH HHS/United States
- T32 DA007315/DA/NIDA NIH HHS/United States
- P42 ES010337/ES/NIEHS NIH HHS/United States
- F32 HD070579/HD/NICHD NIH HHS/United States
- P30 CA023100/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases