Discovery and validation of Barrett's esophagus microRNA transcriptome by next generation sequencing - PubMed (original) (raw)
Discovery and validation of Barrett's esophagus microRNA transcriptome by next generation sequencing
Ajay Bansal et al. PLoS One. 2013.
Abstract
Objective: Barrett's esophagus (BE) is transition from squamous to columnar mucosa as a result of gastroesophageal reflux disease (GERD). The role of microRNA during this transition has not been systematically studied.
Design: For initial screening, total RNA from 5 GERD and 6 BE patients was size fractionated. RNA <70 nucleotides was subjected to SOLiD 3 library preparation and next generation sequencing (NGS). Bioinformatics analysis was performed using R package "DEseq". A p value<0.05 adjusted for a false discovery rate of 5% was considered significant. NGS-identified miRNA were validated using qRT-PCR in an independent group of 40 GERD and 27 BE patients. MicroRNA expression of human BE tissues was also compared with three BE cell lines.
Results: NGS detected 19.6 million raw reads per sample. 53.1% of filtered reads mapped to miRBase version 18. NGS analysis followed by qRT-PCR validation found 10 differentially expressed miRNA; several are novel (-708-5p, -944, -224-5p and -3065-5p). Up- or down- regulation predicted by NGS was matched by qRT-PCR in every case. Human BE tissues and BE cell lines showed a high degree of concordance (70-80%) in miRNA expression. Prediction analysis identified targets that mapped to developmental signaling pathways such as TGFβ and Notch and inflammatory pathways such as toll-like receptor signaling and TGFβ. Cluster analysis found similarly regulated (up or down) miRNA to share common targets suggesting coordination between miRNA.
Conclusion: Using highly sensitive next-generation sequencing, we have performed a comprehensive genome wide analysis of microRNA in BE and GERD patients. Differentially expressed miRNA between BE and GERD have been further validated. Expression of miRNA between BE human tissues and BE cell lines are highly correlated. These miRNA should be studied in biological models to further understand BE development.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Flowchart depicting sequential mapping of the reads.
As demonstrated, the unmapped reads were remapped to miRbase after relaxing the criteria to allow 2–3 mismatches leaving only ∼1% of the reads unmapped. fRNAdb, functional RNA database version 3.4; ‘ambiguous’ represents those reads that mapped to multiple different non-coding RNA in the fRNAdb; ‘others’ includes unclassified ncRNAs in fRNAdb; * these miRNA were not included in the final analysis of differential expression.
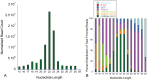
Figure 2. Normalized read counts and their distribution according to the nucleotide length.
Fig. 1A shows that majority of trimmed reads were 21–23 nucleotides in length, the same size as miRNA. Fig. 1B shows the distribution of trimmed reads based on their mapping to miRNA, human genome, non-coding RNA (besides miRNA etc) and E coli genome. mm1, mm2 and mm3 represent alignment to miRBase with 0 or 1, 2 and 3 mismatches respectively. Note that the majority of aligned miRNA with 0 or 1 mismatch are distributed around 22 base pairs, the expected size of miRNA. hg19, human genome version 19; fRNAdb, functional RNA database.
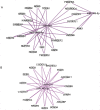
Figure 3. Prediction of target genes for similarly regulated miRNA.
Note that miR -3065, -944 and -149-5p all were down-regulated and share multiple common targets (Panel A) and miR-192 and -215 all were up-regulated and share common targets (Panel B). These results of common targets for similarly up- or down- regulated miRNA suggest coordination between miRNA.
Similar articles
- A detailed analysis of next generation sequencing reads of microRNA expression in Barrett's esophagus: absolute versus relative quantification.
Lee IH, Hong X, Mathur SC, Sharma M, Rastogi A, Sharma P, Christenson LK, Bansal A. Lee IH, et al. BMC Res Notes. 2014 Apr 4;7:212. doi: 10.1186/1756-0500-7-212. BMC Res Notes. 2014. PMID: 24708854 Free PMC article. - Differential MicroRNA Signatures in the Pathogenesis of Barrett's Esophagus.
Craig MP, Rajakaruna S, Paliy O, Sajjad M, Madhavan S, Reddy N, Zhang J, Bottomley M, Agrawal S, Kadakia MP. Craig MP, et al. Clin Transl Gastroenterol. 2020 Jan;11(1):e00125. doi: 10.14309/ctg.0000000000000125. Clin Transl Gastroenterol. 2020. PMID: 31934893 Free PMC article. - MicroRNA profile in neosquamous esophageal mucosa following ablation of Barrett's esophagus.
Sreedharan L, Mayne GC, Watson DI, Bright T, Lord RV, Ansar A, Wang T, Kist J, Astill DS, Hussey DJ. Sreedharan L, et al. World J Gastroenterol. 2017 Aug 14;23(30):5508-5518. doi: 10.3748/wjg.v23.i30.5508. World J Gastroenterol. 2017. PMID: 28852310 Free PMC article. - The biopsy diagnosis of gastroesophageal reflux disease, "carditis," and Barrett's esophagus, and sequelae of therapy.
Riddell RH. Riddell RH. Am J Surg Pathol. 1996;20 Suppl 1:S31-50. doi: 10.1097/00000478-199600001-00005. Am J Surg Pathol. 1996. PMID: 8694147 Review. - MicroRNA dysregulation and therapeutic opportunities in esophageal diseases.
Markey GE, Donohoe CL, McNamee EN, Masterson JC. Markey GE, et al. Am J Physiol Gastrointest Liver Physiol. 2023 Jul 1;325(1):G1-G13. doi: 10.1152/ajpgi.00184.2022. Epub 2023 May 2. Am J Physiol Gastrointest Liver Physiol. 2023. PMID: 37129237 Review.
Cited by
- A detailed analysis of next generation sequencing reads of microRNA expression in Barrett's esophagus: absolute versus relative quantification.
Lee IH, Hong X, Mathur SC, Sharma M, Rastogi A, Sharma P, Christenson LK, Bansal A. Lee IH, et al. BMC Res Notes. 2014 Apr 4;7:212. doi: 10.1186/1756-0500-7-212. BMC Res Notes. 2014. PMID: 24708854 Free PMC article. - A Systematic Review of Esophageal MicroRNA Markers for Diagnosis and Monitoring of Barrett's Esophagus.
Mallick R, Patnaik SK, Wani S, Bansal A. Mallick R, et al. Dig Dis Sci. 2016 Apr;61(4):1039-50. doi: 10.1007/s10620-015-3959-3. Epub 2015 Nov 14. Dig Dis Sci. 2016. PMID: 26572780 Review. - MicroRNA Expression can be a Promising Strategy for the Detection of Barrett's Esophagus: A Pilot Study.
Bansal A, Hong X, Lee IH, Krishnadath KK, Mathur SC, Gunewardena S, Rastogi A, Sharma P, Christenson LK. Bansal A, et al. Clin Transl Gastroenterol. 2014 Dec 11;5(12):e65. doi: 10.1038/ctg.2014.17. Clin Transl Gastroenterol. 2014. PMID: 25502391 Free PMC article. - MiRNA-Related SNPs and Risk of Esophageal Adenocarcinoma and Barrett's Esophagus: Post Genome-Wide Association Analysis in the BEACON Consortium.
Buas MF, Onstad L, Levine DM, Risch HA, Chow WH, Liu G, Fitzgerald RC, Bernstein L, Ye W, Bird NC, Romero Y, Casson AG, Corley DA, Shaheen NJ, Wu AH, Gammon MD, Reid BJ, Hardie LJ, Peters U, Whiteman DC, Vaughan TL. Buas MF, et al. PLoS One. 2015 Jun 3;10(6):e0128617. doi: 10.1371/journal.pone.0128617. eCollection 2015. PLoS One. 2015. PMID: 26039359 Free PMC article. - Barrett's esophagus and cancer risk: how research advances can impact clinical practice.
di Pietro M, Alzoubaidi D, Fitzgerald RC. di Pietro M, et al. Gut Liver. 2014 Jul;8(4):356-70. doi: 10.5009/gnl.2014.8.4.356. Epub 2014 Jul 1. Gut Liver. 2014. PMID: 25071900 Free PMC article. Review.
References
- Eloubeidi MA, Mason AC, Desmond RA, El-Serag HB (2003) Temporal trends (1973–1997) in survival of patients with esophageal adenocarcinoma in the United States: a glimmer of hope? Am J Gastroenterol 98: 1627–1633. - PubMed
- Ness-Jensen E, Lindam A, Lagergren J, Hveem K (2011) Changes in prevalence, incidence and spontaneous loss of gastro-oesophageal reflux symptoms: a prospective population-based cohort study, the HUNT study. Gut - PubMed
- Pohl H, Welch HG (2005) The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst 97: 142–146. - PubMed
- Souza RF, Krishnan K, Spechler SJ (2008) Acid, bile, and CDX: the ABCs of making Barrett's metaplasia. Am J Physiol Gastrointest Liver Physiol 295: G211–218. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical