Epigenetic mechanisms of drug addiction - PubMed (original) (raw)
Review
Epigenetic mechanisms of drug addiction
Jian Feng et al. Curr Opin Neurobiol. 2013 Aug.
Abstract
Epigenetic regulation can mediate long-lasting changes in gene expression, which makes it an attractive mechanism for the stable behavioral abnormalities that characterize drug addiction. Recent research has unveiled numerous types of epigenetic modifications within the brain's reward circuitry in animal models of drug addiction. In this review, we summarize the latest advances in the field, focusing on histone modifications, DNA methylation, and noncoding RNAs. We also highlight several areas for future research. Unraveling the highly complex epigenetic mechanisms of addiction is adding to our understanding of this syndrome and has the potential to trigger novel approaches for better diagnosis and therapy.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
Figure 1. Histone posttranslational modifications
(A) The nucleosome core particle composed of 147 bp of DNA wrapped around an octamer of histone proteins (two copies each of H2A, H2B, H3, and H4). (B) Histone modifications on histone H3 tail. Permissive gene expression correlates with modifications that weaken the interaction between histones and DNA or that promote the recruitment of transcriptional activating complexes (e.g., histone acetylation at K23, K18, K14, and K9, as well as methylation at K79, K36, and K4 or phosphorylation at S28 and S10). Repressive transcription correlates with histone deacetylation (which compacts nucleosomes), histone methylation (e.g., on H3K27 or H3K9, which recruits repressive complexes to chromatin), or DNA methylation (not shown).
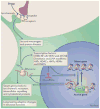
Figure 2. Mechanisms of transcriptional and epigenetic regulation by drugs of abuse
Drugs of abuse act through synaptic targets (reuptake mechanisms, ion channels, and neurotransmitter [NT] receptors) to alter intracellular signalling cascades. This leads to the activation or inhibition of transcription factors and of many other nuclear targets, including chromatin-regulatory proteins (shown by thick arrows). These processes result in the induction or repression of particular genes, which can in turn further regulate gene transcription. It is proposed that some of these drug-induced changes at the chromatin level are extremely stable and thereby underlie the long-lasting behaviours that define addiction. CREB, cAMP-response element binding protein; DNMTs, DNA methyltransferases; HATs, histone acetyltransferases; HDACs, histone deacetylases; HDMs, histone demethylases; HMTs, histone methyltransferases; MEF2, myocyte-specific enhancer factor 2; NF-κB, nuclear factor-κB; pol II, RNA polymerase II. Figure is reproduced with permission from Ref.
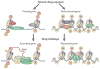
Figure 3. Gene priming and desensitization
Epigenetic mechanisms are important in mediating gene priming and desensitization, which are not reflected by stable changes in steady-state mRNA levels. Instead, a later drug challenge induces a given gene to a greater (primed) or lesser (desensitized) extent based on the epigenetic modifications that are induced by previous chronic drug exposure. A, acetylation; M, methylation; P, phosphorylation; pol II, RNA polymerase II. Figure is reproduced with permission from Ref. .
Similar articles
- Epigenetic Mechanisms of Psychostimulant-Induced Addiction.
Kalda A, Zharkovsky A. Kalda A, et al. Int Rev Neurobiol. 2015;120:85-105. doi: 10.1016/bs.irn.2015.02.010. Epub 2015 Apr 8. Int Rev Neurobiol. 2015. PMID: 26070754 Review. - Neuroepigenetics and addiction.
Walker DM, Nestler EJ. Walker DM, et al. Handb Clin Neurol. 2018;148:747-765. doi: 10.1016/B978-0-444-64076-5.00048-X. Handb Clin Neurol. 2018. PMID: 29478612 Free PMC article. Review. - Epigenetic mechanisms of drug addiction.
Nestler EJ. Nestler EJ. Neuropharmacology. 2014 Jan;76 Pt B(0 0):259-68. doi: 10.1016/j.neuropharm.2013.04.004. Epub 2013 Apr 30. Neuropharmacology. 2014. PMID: 23643695 Free PMC article. Review. - Epigenetics and addiction.
Hamilton PJ, Nestler EJ. Hamilton PJ, et al. Curr Opin Neurobiol. 2019 Dec;59:128-136. doi: 10.1016/j.conb.2019.05.005. Epub 2019 Jun 27. Curr Opin Neurobiol. 2019. PMID: 31255844 Free PMC article. Review. - Mechanisms of epigenetic memory and addiction.
Tuesta LM, Zhang Y. Tuesta LM, et al. EMBO J. 2014 May 16;33(10):1091-103. doi: 10.1002/embj.201488106. Epub 2014 Apr 28. EMBO J. 2014. PMID: 24778453 Free PMC article. Review.
Cited by
- Three-dimensional chromosome architecture and drug addiction.
Chitaman JM, Fraser P, Feng J. Chitaman JM, et al. Curr Opin Neurobiol. 2019 Dec;59:137-145. doi: 10.1016/j.conb.2019.05.009. Epub 2019 Jul 2. Curr Opin Neurobiol. 2019. PMID: 31276935 Free PMC article. Review. - How Preclinical Models Evolved to Resemble the Diagnostic Criteria of Drug Addiction.
Belin-Rauscent A, Fouyssac M, Bonci A, Belin D. Belin-Rauscent A, et al. Biol Psychiatry. 2016 Jan 1;79(1):39-46. doi: 10.1016/j.biopsych.2015.01.004. Epub 2015 Jan 29. Biol Psychiatry. 2016. PMID: 25747744 Free PMC article. Review. - A multiancestry study identifies novel genetic associations with CHRNA5 methylation in human brain and risk of nicotine dependence.
Hancock DB, Wang JC, Gaddis NC, Levy JL, Saccone NL, Stitzel JA, Goate A, Bierut LJ, Johnson EO. Hancock DB, et al. Hum Mol Genet. 2015 Oct 15;24(20):5940-54. doi: 10.1093/hmg/ddv303. Epub 2015 Jul 28. Hum Mol Genet. 2015. PMID: 26220977 Free PMC article. - Investigating the potential influence of cause of death and cocaine levels on the differential expression of genes associated with cocaine abuse.
Bannon MJ, Savonen CL, Hartley ZJ, Johnson MM, Schmidt CJ. Bannon MJ, et al. PLoS One. 2015 Feb 6;10(2):e0117580. doi: 10.1371/journal.pone.0117580. eCollection 2015. PLoS One. 2015. PMID: 25658879 Free PMC article. - Long non-coding RNA MEG3 attends to morphine-mediated autophagy of HT22 cells through modulating ERK pathway.
Gao S, Li E, Gao H. Gao S, et al. Pharm Biol. 2019 Dec;57(1):536-542. doi: 10.1080/13880209.2019.1651343. Pharm Biol. 2019. PMID: 31433241 Free PMC article.
References
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. - PubMed
- Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nat Rev Neurosci. 2001;2:119–128. - PubMed
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical