Role of the gut microbiota in the development and function of lymphoid cells - PubMed (original) (raw)
Review
Role of the gut microbiota in the development and function of lymphoid cells
Nobuhiko Kamada et al. J Immunol. 2013.
Abstract
Mammals are colonized by large numbers of microorganisms, including trillions of bacteria, most of which live in the intestinal tract. These indigenous microorganisms that inhabit the body of humans and animals are referred collectively to as the microbiota. Accumulating evidence indicates that the microbiota regulates the development and/or function of different types of immune cells in the intestine. For example, the microbiota drives homeostatic, pathogenic, and regulatory T cell immune responses that contribute to tissue homeostasis, but also can promote disease. The gut microbes also facilitate IgA responses, which in turn regulate the composition and function of the gut microbiota. Thus, the reciprocal regulation of the gut microbiota and the host immune system may influence the balance between homeostasis and disease in the intestine.
Conflict of interest statement
Disclosures:
The authors have no conflicting financial interests.
Figures
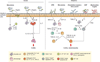
Figure 1. Gut microbiota drive homeostatic, pathogenic and regulatory lymphocyte immunity
The gut microbiota directly or indirectly regulates the function of TCRγδ T cells. In turn, TCRγδ T cells regulate the microbiota through induction of the anti-microbial peptide RegIIIγ. The microbiota positively or negatively regulate the differentiation and/or function of RORγt+ ILCs. The microbiota stabilize the expression of RORγt+ in ILCs through induction of IL-7. The absence of microbiota-induced IL-7 signaling or the presence of IL-12 and IL-15 facilitates the conversion of RORγt+ ILCs into RORγt− IFN-γ-producing pathogenic ILCs. Segmented filamentous bacteria (SFB) promote the differentiation of Th17 cells in the gut. Steady-state Th17 cells produce IL-17, IL-22 and IL-10 and may play an homeostatic role in the gut. IL-23, IL-12, IL-1β, TGF-β3 induce conversion of homeostatic Th17 cells towards pathogenic, IFN-γ-producing Th17 cells that facilitate intestinal inflammation. Certain subsets of commensal microbiota, such as Clostridium clusters XIVa and IV and Bacteroides fragilis, promote the development and functional maturation of Foxp3+Helios− inducible Tregs. Foxp3+ iTregs suppress the expansion of Th17 cells as well as their conversion into IFN-γ-producing Th17 cells.
Similar articles
- The Role of Microbiota on the Gut Immunology.
Min YW, Rhee PL. Min YW, et al. Clin Ther. 2015 May 1;37(5):968-75. doi: 10.1016/j.clinthera.2015.03.009. Epub 2015 Apr 4. Clin Ther. 2015. PMID: 25846321 Review. - Intestinal IgA synthesis: a primitive form of adaptive immunity that regulates microbial communities in the gut.
Suzuki K, Ha SA, Tsuji M, Fagarasan S. Suzuki K, et al. Semin Immunol. 2007 Apr;19(2):127-35. doi: 10.1016/j.smim.2006.10.001. Epub 2006 Dec 11. Semin Immunol. 2007. PMID: 17161619 Review. - Intestinal IgA production and its role in host-microbe interaction.
Gutzeit C, Magri G, Cerutti A. Gutzeit C, et al. Immunol Rev. 2014 Jul;260(1):76-85. doi: 10.1111/imr.12189. Immunol Rev. 2014. PMID: 24942683 Free PMC article. Review. - Effective Barriers: The Role of NKT Cells and Innate Lymphoid Cells in the Gut.
Cairo C, Webb TJ. Cairo C, et al. J Immunol. 2022 Jan 15;208(2):235-246. doi: 10.4049/jimmunol.2100799. J Immunol. 2022. PMID: 35017213 Review. - Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease.
Pickard JM, Zeng MY, Caruso R, Núñez G. Pickard JM, et al. Immunol Rev. 2017 Sep;279(1):70-89. doi: 10.1111/imr.12567. Immunol Rev. 2017. PMID: 28856738 Free PMC article. Review.
Cited by
- Sodium Chloride Aggravates Arthritis via Th17 Polarization.
Jung SM, Kim Y, Kim J, Jung H, Yi H, Rim YA, Park N, Kwok SK, Park SH, Ju JH. Jung SM, et al. Yonsei Med J. 2019 Jan;60(1):88-97. doi: 10.3349/ymj.2019.60.1.88. Yonsei Med J. 2019. PMID: 30554495 Free PMC article. - Contribution of the Commensal Microflora to the Immunological Homeostasis and the Importance of Immune-Related Drug Development for Clinical Applications.
Beutgen VM, Schmelter C, Pfeiffer N, Grus FH. Beutgen VM, et al. Int J Mol Sci. 2021 Aug 18;22(16):8896. doi: 10.3390/ijms22168896. Int J Mol Sci. 2021. PMID: 34445599 Free PMC article. Review. - Intestinal microbiota imbalance resulted by anti-Toxoplasma gondii immune responses aggravate gut and brain injury.
Chen J, Zhang C, Yang Z, Wu W, Zou W, Xin Z, Zheng S, Liu R, Yang L, Peng H. Chen J, et al. Parasit Vectors. 2024 Jul 2;17(1):284. doi: 10.1186/s13071-024-06349-8. Parasit Vectors. 2024. PMID: 38956725 Free PMC article. - Anti-Inflammatory and Immunomodulatory Properties of Fermented Plant Foods.
Shahbazi R, Sharifzad F, Bagheri R, Alsadi N, Yasavoli-Sharahi H, Matar C. Shahbazi R, et al. Nutrients. 2021 Apr 30;13(5):1516. doi: 10.3390/nu13051516. Nutrients. 2021. PMID: 33946303 Free PMC article. Review. - A systematic review of enteric dysbiosis in chronic fatigue syndrome/myalgic encephalomyelitis.
Du Preez S, Corbitt M, Cabanas H, Eaton N, Staines D, Marshall-Gradisnik S. Du Preez S, et al. Syst Rev. 2018 Dec 20;7(1):241. doi: 10.1186/s13643-018-0909-0. Syst Rev. 2018. PMID: 30572962 Free PMC article.
References
- Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. - PubMed
- Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 2010;140:845–858. - PubMed
- Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, Takeda K. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK091191/DK/NIDDK NIH HHS/United States
- R01 DK61707/DK/NIDDK NIH HHS/United States
- R01 DK061707/DK/NIDDK NIH HHS/United States
- R01 AR059688/AR/NIAMS NIH HHS/United States
- R01 DK095782/DK/NIDDK NIH HHS/United States
- R01 AI063331/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous