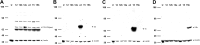Systematic analysis of rat 12/15-lipoxygenase enzymes reveals critical role for spinal eLOX3 hepoxilin synthase activity in inflammatory hyperalgesia - PubMed (original) (raw)
Systematic analysis of rat 12/15-lipoxygenase enzymes reveals critical role for spinal eLOX3 hepoxilin synthase activity in inflammatory hyperalgesia
Ann M Gregus et al. FASEB J. 2013 May.
Abstract
Previously, we observed significant increases in spinal 12-lipoxygenase (LOX) metabolites, in particular, hepoxilins, which contribute to peripheral inflammation-induced tactile allodynia. However, the enzymatic sources of hepoxilin synthase (HXS) activity in rats remain elusive. Therefore, we overexpressed each of the 6 rat 12/15-LOX enzymes in HEK-293T cells and measured by LC-MS/MS the formation of HXB3, 12-HETE, 8-HETE, and 15-HETE from arachidonic acid (AA) at baseline and in the presence of LOX inhibitors (NDGA, AA-861, CDC, baicalein, and PD146176) vs. vehicle-treated and mock-transfected controls. We detected the following primary intrinsic activities: 12-LOX (Alox12, Alox15), 15-LOX (Alox15b), and HXS (Alox12, Alox15). Similar to human and mouse orthologs, proteins encoded by rat Alox12b and Alox12e possessed minimal 12-LOX activity with AA as substrate, while eLOX3 (encoded by Aloxe3) exhibited HXS without 12-LOX activity when coexpressed with Alox12b or supplemented with 12-HpETE. CDC potently inhibited HXS and 12-LOX activity in vitro (relative IC50s: CDC, ~0.5 and 0.8 μM, respectively) and carrageenan-evoked tactile allodynia in vivo. Notably, peripheral inflammation significantly increased spinal eLOX3; intrathecal pretreatment with either siRNA targeting Aloxe3 or an eLOX3-selective antibody attenuated the associated allodynia. These findings implicate spinal eLOX3-mediated hepoxilin synthesis in inflammatory hyperesthesia and underscore the importance of developing more selective 12-LOX/HXS inhibitors.
Figures
Figure 1.
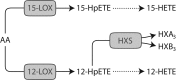
Putative activities of the rat 12/15-LOX enzyme family using arachidonic acid (AA) as substrate. AA can be metabolized by each of 6 rat 12-LOX family enzymes (see Table 1), which exhibit one or more of the following activities depicted above: 15-LOX, 12-LOX, or HXS.
Figure 2.
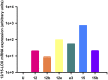
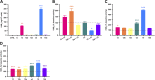
Experimental validation of rat 12/15-LOX overexpression systems. mRNA expression of rat 12/15-LOX enzymes overexpressed in HEK-293T cells (HEK-12/15-LOX) as detected by qPCR. Transient transfection: U, untransfected control; 12, Alox12; 12b, Alox12b; 12e, Alox12e; e3, Aloxe3; 15, Alox15; 15b, Alox15b.
Figure 3.
Experimental validation of rat 12/15-LOX antibodies. Antibodies detecting all six 12/15-LOX isozymes 12-LOX (pan; A) or those selectively targeting e3 (Aloxe3; B), 15 (Alox15; C), or 15b (Alox15b; D), as revealed by immunoblot of lysates from HEK-293T cells overexpressing each of the 6 rat 12/15-LOX enzymes. Samples were run with β-actin as a loading control in each lane as follows: U, untransfected control; 12, Alox12; 12b, Alox12b; 12e, Alox12e; e3, Aloxe3; 15, Alox15; 15b, Alox15b.
Figure 4.
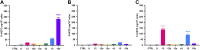
Activities of overexpressed rat 12/15-LOX enzymes. 15-LOX, 8-LOX, or 12-LOX activity with exposure of HEK-12/15-LOX cells to AA substrate as revealed by the release of 15-HETE (A), 8-HETE (B), or 12-HETE (C) from HEK-293T cells either untreated (ctrl) or treated with AA (70 μM), as follows: U, untransfected control; 12, Alox12; 12b, Alox12b; 12e, Alox12e; e3, Aloxe3; 15, Alox15; 15b, Alox15b. ***P < 0.001 vs. untransfected control; n = 4 wells/treatment.
Figure 5.
Formation of hepoxilins observed from rat 12(S)- and (R)-type LOXs and Aloxe3/eLOX3 using AA or 12-HpETE as substrate. Hepoxilins (HXB3) are synthesized directly from AA (70 μM) by Alox12 or Alox15 (A); by Aloxe3 when it is coexpressed at a 1:1 ratio with Alox12b (B); from 12(S)-HpETE (5 μM) by Alox12, Aloxe3, or Alox15 (C); or from racemic 12(R,S)-HpETE (10 μM; D). U, untransfected control; 12, Alox12; 12b, Alox12b; 12e, Alox12e; e3, Aloxe3; 15, Alox15; 15b, Alox15b. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control or baseline; n = 4 wells/treatment.
Figure 6.
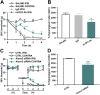
Spinal expression of the hepoxilin synthase eLOX3 is increased following peripheral inflammation. Quantitative PCR and immunoblot reveal isozyme-specific expression of 12/15-LOX in rat spinal cord. Basal expression of Alox12, Alox12b, Aloxe3, and Alox15 mRNA (A) and induction of 12/15-LOX (as detected by pan antibody; B) and eLOX3 (Aloxe3; C) but not 12/15-LOX-l (Alox15) proteins (D) in spinal cord at 4 h after IPLT carrageenan. U, untreated/naive; C, 4 h postintraplantar carrageenan; +, transfected HEK293T cell lysate positive controls for Alox12 (B), Aloxe3 (C), or Alox15 (D). Total protein loaded: spinal cord, 30 μg; positive control, 10 μg. *P < 0.05 vs. control (untreated); n = 6 rats/group.
Figure 7.
Spinal inhibition of eLOX3 or knockdown of Aloxe3 attenuates inflammatory hyperesthesia. IT pretreatment with either prevalidated antibody (5 μg) selectively targeting eLOX3 protein encoded by Aloxe3 (e3; A, B) or siRNA (2 μg; C, D) against Aloxe3 significantly reduced carrageenan (carra)-induced tactile allodynia. *P < 0.05, **P < 0.01 vs. controls (saline vehicle, 5 μg rabbit IgG, or 2 μg nontargeting siRNA); n = 6–10 rats/group.
Similar articles
- Spinal 12-lipoxygenase-derived hepoxilin A3 contributes to inflammatory hyperalgesia via activation of TRPV1 and TRPA1 receptors.
Gregus AM, Doolen S, Dumlao DS, Buczynski MW, Takasusuki T, Fitzsimmons BL, Hua XY, Taylor BK, Dennis EA, Yaksh TL. Gregus AM, et al. Proc Natl Acad Sci U S A. 2012 Apr 24;109(17):6721-6. doi: 10.1073/pnas.1110460109. Epub 2012 Apr 9. Proc Natl Acad Sci U S A. 2012. PMID: 22493235 Free PMC article. - The rat leukocyte-type 12-lipoxygenase exhibits an intrinsic hepoxilin A3 synthase activity.
Nigam S, Patabhiraman S, Ciccoli R, Ishdorj G, Schwarz K, Petrucev B, Kühn H, Haeggström JZ. Nigam S, et al. J Biol Chem. 2004 Jul 9;279(28):29023-30. doi: 10.1074/jbc.M307576200. Epub 2004 May 3. J Biol Chem. 2004. PMID: 15123652 - Mutations associated with a congenital form of ichthyosis (NCIE) inactivate the epidermal lipoxygenases 12R-LOX and eLOX3.
Yu Z, Schneider C, Boeglin WE, Brash AR. Yu Z, et al. Biochim Biophys Acta. 2005 Jan 5;1686(3):238-47. doi: 10.1016/j.bbalip.2004.10.007. Biochim Biophys Acta. 2005. PMID: 15629692 - Hepoxilin A3 synthase.
Nigam S, Zafiriou MP. Nigam S, et al. Biochem Biophys Res Commun. 2005 Dec 9;338(1):161-8. doi: 10.1016/j.bbrc.2005.09.065. Epub 2005 Sep 21. Biochem Biophys Res Commun. 2005. PMID: 16198304 Review. - Structure, biochemistry and biology of hepoxilins: an update.
Nigam S, Zafiriou MP, Deva R, Ciccoli R, Roux-Van der Merwe R. Nigam S, et al. FEBS J. 2007 Jul;274(14):3503-3512. doi: 10.1111/j.1742-4658.2007.05910.x. Epub 2007 Jul 2. FEBS J. 2007. PMID: 17608719 Review.
Cited by
- Role of leukotriene B4 (LTB4)-LTB4 receptor 1 signaling in post-incisional nociceptive sensitization and local inflammation in mice.
Asahara M, Ito N, Hoshino Y, Sasaki T, Yokomizo T, Nakamura M, Shimizu T, Yamada Y. Asahara M, et al. PLoS One. 2022 Oct 20;17(10):e0276135. doi: 10.1371/journal.pone.0276135. eCollection 2022. PLoS One. 2022. PMID: 36264904 Free PMC article. - 15-Lipoxygenases regulate the production of chemokines in human lung macrophages.
Abrial C, Grassin-Delyle S, Salvator H, Brollo M, Naline E, Devillier P. Abrial C, et al. Br J Pharmacol. 2015 Sep;172(17):4319-30. doi: 10.1111/bph.13210. Epub 2015 Jul 14. Br J Pharmacol. 2015. PMID: 26040494 Free PMC article. - An ALOX12-12-HETE-GPR31 signaling axis is a key mediator of hepatic ischemia-reperfusion injury.
Zhang XJ, Cheng X, Yan ZZ, Fang J, Wang X, Wang W, Liu ZY, Shen LJ, Zhang P, Wang PX, Liao R, Ji YX, Wang JY, Tian S, Zhu XY, Zhang Y, Tian RF, Wang L, Ma XL, Huang Z, She ZG, Li H. Zhang XJ, et al. Nat Med. 2018 Jan;24(1):73-83. doi: 10.1038/nm.4451. Epub 2017 Dec 11. Nat Med. 2018. PMID: 29227475 - Ferroptosis: mechanisms and links with diseases.
Yan HF, Zou T, Tuo QZ, Xu S, Li H, Belaidi AA, Lei P. Yan HF, et al. Signal Transduct Target Ther. 2021 Feb 3;6(1):49. doi: 10.1038/s41392-020-00428-9. Signal Transduct Target Ther. 2021. PMID: 33536413 Free PMC article. Review. - The absence of the leukotriene B4 receptor BLT1 attenuates peripheral inflammation and spinal nociceptive processing following intraplantar formalin injury.
Asahara M, Ito N, Yokomizo T, Nakamura M, Shimizu T, Yamada Y. Asahara M, et al. Mol Pain. 2015 Mar 12;11:11. doi: 10.1186/s12990-015-0010-9. Mol Pain. 2015. PMID: 25889478 Free PMC article.
References
- Kuhn H., O'Donnell V. B. (2006) Inflammation and immune regulation by 12/15-lipoxygenases. Prog. Lipid Res. 45, 334–356 - PubMed
- Ivanov I., Heydeck D., Hofheinz K., Roffeis J., O'Donnell V. B., Kuhn H., Walther M. (2010) Molecular enzymology of lipoxygenases. Arch. Biochem. Biophys. 503, 161–174 - PubMed
- Krieg P., Marks F., Furstenberger G. (2001) A gene cluster encoding human epidermis-type lipoxygenases at chromosome 17p13.1: cloning, physical mapping, and expression. Genomics 73, 323–330 - PubMed
- Bendani M. K., Palluy O., Cook-Moreau J., Beneytout J. L., Rigaud M., Vallat J. M. (1995) Localization of 12-lipoxygenase mRNA in cultured oligodendrocytes and astrocytes by in situ reverse transcriptase and polymerase chain reaction. Neurosci. Lett. 189, 159–162 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DA002110/DA/NIDA NIH HHS/United States
- U54 GM069338/GM/NIGMS NIH HHS/United States
- NS16541/NS/NINDS NIH HHS/United States
- R37 NS016541/NS/NINDS NIH HHS/United States
- R01 NS016541/NS/NINDS NIH HHS/United States
- T32 GM007752/GM/NIGMS NIH HHS/United States
- GM064611/GM/NIGMS NIH HHS/United States
- GM069338/GM/NIGMS NIH HHS/United States
- DA02110/DA/NIDA NIH HHS/United States
- R01 GM064611/GM/NIGMS NIH HHS/United States
- T32 DK007202/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous