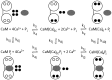Calmodulin transduces Ca2+ oscillations into differential regulation of its target proteins - PubMed (original) (raw)
. 2013 Apr 17;4(4):601-12.
doi: 10.1021/cn300218d. Epub 2013 Feb 5.
Affiliations
- PMID: 23384199
- PMCID: PMC3629746
- DOI: 10.1021/cn300218d
Calmodulin transduces Ca2+ oscillations into differential regulation of its target proteins
Nikolai Slavov et al. ACS Chem Neurosci. 2013.
Abstract
Diverse physiological processes are regulated differentially by Ca(2+) oscillations through the common regulatory hub calmodulin. The capacity of calmodulin to combine specificity with promiscuity remains to be resolved. Here we propose a mechanism based on the molecular properties of calmodulin, its two domains with separate Ca(2+) binding affinities, and target exchange rates that depend on both target identity and Ca(2+) occupancy. The binding dynamics among Ca(2+), Mg(2+), calmodulin, and its targets were modeled with mass-action differential equations based on experimentally determined protein concentrations and rate constants. The model predicts that the activation of calcineurin and nitric oxide synthase depends nonmonotonically on Ca(2+)-oscillation frequency. Preferential activation reaches a maximum at a target-specific frequency. Differential activation arises from the accumulation of inactive calmodulin-target intermediate complexes between Ca(2+) transients. Their accumulation provides the system with hysteresis and favors activation of some targets at the expense of others. The generality of this result was tested by simulating 60 000 networks with two, four, or eight targets with concentrations and rate constants from experimentally determined ranges. Most networks exhibit differential activation that increases in magnitude with the number of targets. Moreover, differential activation increases with decreasing calmodulin concentration due to competition among targets. The results rationalize calmodulin signaling in terms of the network topology and the molecular properties of calmodulin.
Figures
Figure 1
Calmodulin. (A) NMR structure of apo calmodulin (1cfc.pdb), (B) X-ray structure of CaM-Ca4 (1cll.pdb) and (C) crystal structure of CaM-Ca4 peptide complex (1cdl.pdb). Panels A–C were prepared using MOLMOL with EF1 in light green, EF2 in green, EF3 in light blue, and EF4 in blue. Ca2+ ions are shown as black balls and the target peptide in red. (D) Fraction of calmodulin with one (light blue, dashed), two (blue), three (light green, dashed), and four (green) Ca2+ bound as a function of free Ca2+ concentration at physiological KCl concentration based on experimental binding constants.
Figure 2
Network model. Reactions and species taken into account in the simulations are shown. Calmodulin (CaM) is shown as a dumbbell indicating its two-domain structure, open circles denote unoccupied Ca2+-binding sites, filled black circles are Ca2+ ions, and the gray oval denotes the _j_th target protein. The subscript 2C indicates two Ca2+ ions in the C-terminal domain of calmodulin, and subscript j indicates the _j_th target protein. The top line cartoon and reactions indicate stepwise reversible binding of pairs of Ca2+ ions to calmodulin. The bottom line cartoon and reactions indicate stepwise reversible binding of pairs of Ca2+ ions to calmodulin in complex with the _j_th target protein. The vertical reactions indicate reversible binding of the _j_th target.
Figure 3
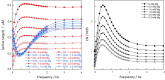
Activation of CN and NOS by Ca2+ oscillations. Concentrations of network species are shown as a function of time under different simulation conditions. The Mg2+ concentration and the midheight duration of Ca2+ transients are given above each set of panels (A–D). Within each set of panels, the top panels show active species with CaM(Ca)4CN in red and CaM(Ca)4NOS in blue; the middle panels show inactive intermediates with CaM(Ca)2CCN in magenta, CaM(Ca)2CNOS in green, CaM(Ca)2C(Mg)2NCN in yellow, and CaM(Ca)2C(Mg)2NNOS in cyan. The bottom panels show the concentration of free Ca2+ (black), oscillating at low frequency (0.04 Hz; left) and at high frequency (1 Hz; right).
Figure 4
Frequency dependence of CN and NOS activity. (Left) Concentration of active target CN (CaM(Ca)4CN in red) and NOS (CaM(Ca)4NOS in blue) and (right) ratio of concentrations of active species (CaM(Ca)4CN over CaM(Ca)4NOS) on the 11th transient as a function of Ca2+ frequency. Data at the following Mg2+ concentrations are shown: 0 mM (filled circle), 0.5 mM (filled upward triangle), 1.0 mM (filled diamond), 1.5 mM (filled downward triangle), 2.0 mM (open circle), and 2.5 mM (open triangle).
Figure 5
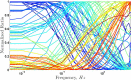
Frequency dependence in generalized networks. Ratio of active targets in a network of two targets using parameters picked randomly within the ranges in Table 2, and calmodulin concentration in the range 1–100 μM. The Ca2+ train consisted of 11 transients with 157 ms midheight duration and simulations were performed at Ca2+ oscillation frequency ranging from 0.005 to 5 Hz. Each curve represents one pair of targets and the different colors are used only to facilitate viewing of the curves.
Figure 6
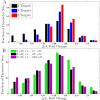
Differential activation in generalized networks. Distribution of Δ_A_ values among 10 000 simulations for generalized networks with rate constants generated at random within the ranges given in Table 2. Δ_A_ is calculated comparing 1 and 0.04 Hz oscillations, and the fractions of simulations yielding the indicated ranges of Δ_A_ values are shown. The simulations used trains of 11 Ca2+ transients with 157 ms midheight duration without Mg2+. (A) Effect of number of targets. Δ_A_ distributions for networks of two (black), four (blue), or eight (red) targets. The calmodulin concentration was chosen randomly in the range 1–100 μM, and the total target concentration in the range 1–100 μM. (B) Effect of limiting calmodulin concentration. Distribution of Δ_A_ values for networks containing two targets and calmodulin concentration sampled in the ranges 1–10 μM (green), 1–100 μM (black), or 1–1000 μM (magenta).
Figure 7
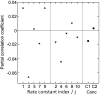
Sensitivity analysis. Partial correlation coefficient between Δ_A_ and each varied parameter for 10 000 simulations of networks having two targets and no magnesium. The rate constants _ki_j (indicated by their indices i) are displayed as upward triangle (association rate constants) or downward triangle (dissociation rate constants) and concentrations as filled circles (C1 = total calmodulin concentration, C2 = target concentration).
Similar articles
- Small proteins that modulate calmodulin-dependent signal transduction: effects of PEP-19, neuromodulin, and neurogranin on enzyme activation and cellular homeostasis.
Slemmon JR, Feng B, Erhardt JA. Slemmon JR, et al. Mol Neurobiol. 2000 Aug-Dec;22(1-3):99-113. doi: 10.1385/MN:22:1-3:099. Mol Neurobiol. 2000. PMID: 11414283 Review. - Dual calcium ion regulation of calcineurin by calmodulin and calcineurin B.
Stemmer PM, Klee CB. Stemmer PM, et al. Biochemistry. 1994 Jun 7;33(22):6859-66. doi: 10.1021/bi00188a015. Biochemistry. 1994. PMID: 8204620 - Differential integration of Ca2+-calmodulin signal in intact ventricular myocytes at low and high affinity Ca2+-calmodulin targets.
Song Q, Saucerman JJ, Bossuyt J, Bers DM. Song Q, et al. J Biol Chem. 2008 Nov 14;283(46):31531-40. doi: 10.1074/jbc.M804902200. Epub 2008 Sep 12. J Biol Chem. 2008. PMID: 18790737 Free PMC article. - Dynamics of nitric oxide synthase-calmodulin interactions at physiological calcium concentrations.
Piazza M, Guillemette JG, Dieckmann T. Piazza M, et al. Biochemistry. 2015 Mar 24;54(11):1989-2000. doi: 10.1021/bi501353s. Epub 2015 Mar 16. Biochemistry. 2015. PMID: 25751535 - The Ca2+/calmodulin signaling system in the neural response to excitability. Involvement of neuronal and glial cells.
Solà C, Barrón S, Tusell JM, Serratosa J. Solà C, et al. Prog Neurobiol. 1999 Jun;58(3):207-32. doi: 10.1016/s0301-0082(98)00082-3. Prog Neurobiol. 1999. PMID: 10341361 Review.
Cited by
- Constant growth rate can be supported by decreasing energy flux and increasing aerobic glycolysis.
Slavov N, Budnik BA, Schwab D, Airoldi EM, van Oudenaarden A. Slavov N, et al. Cell Rep. 2014 May 8;7(3):705-14. doi: 10.1016/j.celrep.2014.03.057. Epub 2014 Apr 24. Cell Rep. 2014. PMID: 24767987 Free PMC article. - Combined Pulsed Electron Double Resonance EPR and Molecular Dynamics Investigations of Calmodulin Suggest Effects of Crowding Agents on Protein Structures.
Stewart AM, Shanmugam M, Kutta RJ, Scrutton NS, Lovett JE, Hay S. Stewart AM, et al. Biochemistry. 2022 Sep 6;61(17):1735-1742. doi: 10.1021/acs.biochem.2c00099. Epub 2022 Aug 18. Biochemistry. 2022. PMID: 35979922 Free PMC article. - Physiologically Relevant Free Ca2+ Ion Concentrations Regulate STRA6-Calmodulin Complex Formation via the BP2 Region of STRA6.
Young BD, Varney KM, Wilder PT, Costabile BK, Pozharski E, Cook ME, Godoy-Ruiz R, Clarke OB, Mancia F, Weber DJ. Young BD, et al. J Mol Biol. 2021 Nov 5;433(22):167272. doi: 10.1016/j.jmb.2021.167272. Epub 2021 Sep 27. J Mol Biol. 2021. PMID: 34592217 Free PMC article. - Competitive tuning: Competition's role in setting the frequency-dependence of Ca2+-dependent proteins.
Romano DR, Pharris MC, Patel NM, Kinzer-Ursem TL. Romano DR, et al. PLoS Comput Biol. 2017 Nov 6;13(11):e1005820. doi: 10.1371/journal.pcbi.1005820. eCollection 2017 Nov. PLoS Comput Biol. 2017. PMID: 29107982 Free PMC article. - Single cell protein analysis for systems biology.
Levy E, Slavov N. Levy E, et al. Essays Biochem. 2018 Oct 26;62(4):595-605. doi: 10.1042/EBC20180014. Print 2018 Oct 26. Essays Biochem. 2018. PMID: 30072488 Free PMC article. Review.
References
- Dolmetsch R. E.; Xu K.; Lewis R. S. (1998) Calcium oscillations increase the efficiency and specificity of gene expression. Nature 392, 933–936. - PubMed
- Berridge M. J. (2012) Calcium signalling remodelling and disease,,. Biochem. Soc. Trans. 40, 297–309. - PubMed
- Berridge M. J.; Bootman M. D.; Roderick H. L. (2003) Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell. Biol. 4, 517–529. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous