Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy - PubMed (original) (raw)
Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy
Sai Li et al. Sci Signal. 2013.
Abstract
Autophagy, the process of lysosome-dependent degradation of cytosolic components, is a mechanism by which cells selectively engulf invading pathogens to protect themselves against infection. Galectin-8, a cytosolic protein with specificity for β-galactoside-containing glycans, binds endosomal and lysosomal membranes that have been damaged, for example, by pathogens, and selectively recruits the autophagy cargo receptor NDP52 to induce autophagy. We solved the crystal structure of the NDP52-galectin-8 complex to show how NDP52 exclusively binds galectin-8 and, consequently, why other galectins do not restrict the growth of Salmonella in human cells.
Figures
Figure 1. The molecular basis of the interaction between Gal8C-CRD and NDP52368-381
(A) Cartoon representation of Gal8C-CRD (yellow and grey) in complex with NDP52368-381 (cyan). F1-F5, S1-S6 numbering of β-strands. (B) Hydrophobic contacts between Gal8C-CRD and NDP52368-381. Residues of Gal8C-CRD involved in hydrophobic interaction are colored yellow and labeled in black. NDP52 in cyan. (C) ITC data for the interaction of NDP52368-381and Gal8C-CRD at the indicated peptide versus protein molar ratios. Top, raw data; bottom, integrated heat data. Kd, dissociation constant; N, reaction stoichiometry. (D) Hydrogen bonds between Gal8C-CRD (yellow) and NDP52368-381 (cyan) with distances measured in Ångströms (Å). (E) Dissociation constants for the indicated interactions obtained through ITC. . NB: binding not detectable. (F) Scheme of interactions between NDP52 (blue) and galectin-8 (yellow). Single letter abbreviations indicate mutations tested in this study with colors indicating affinity (red: Kd >120 μM, blue: 27< Kd <41 μM, black: Kd <12 μM). Circled residues occur in human galectin CRDs, squared residues are experimental mutations.
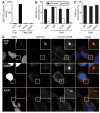
Figure 2. Functional importance of the NDP52 binding site in galectin-8 for anti-bacterial autophagy
(A) LUMIER binding assay. Normalized ratio of luciferase activity bound to anti-Flag beads to luciferase activity in lysates from 293ET cells co-expressing NDP52 fused to luciferase and the indicated Flag-tagged wild-type (WT) or mutated (I312A, A323Y) galectin-8 proteins. (B-C) Percentage of S. Typhimurium exhibiting NDP52 (B) or galectin-8 (C) recruitment at 1 hour post-infection in Hela cells expressing the indicated siRNA-resistant WT or mutated galectin-8 alleles fused to mCherry as determined by fluorescence microscopy. Cells were treated with control siRNA (siControl) or siRNA against galectin-8 (siGal8) (B) or only siGal8 (C).. Mean and s.d. were quantified from quadruplicate cultures (n>200 bacteria per coverslip), and representative confocal images from siGal8-treated cells are shown (D).
Figure 3. The molecular basis for the specificity of NDP52 for galectin-8
(A) For galectins containing two CRDs, N- and C-terminal domains are indicated. Red dots and black triangles indicate residues in Gal8C-CRD that, if mutated, prevented or impaired binding to NDP52368-381, respectively. (B) Close-up view of Gal8 Ala323 (yellow) in contact with the γ-carbon of the NDP52 Pro379 and the ε-carbon and hydroxyl oxygen of the NDP52 Tyr380 side chain (blue and red). Van der Waals volumes of key atoms rendered as transparent spheres. (C) Dissociation constants for the interaction of NDP52368-381 with the indicated Gal8C-CRD variants obtained ITC. NB: binding not detectable.
Similar articles
- Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion.
Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Thurston TL, et al. Nature. 2012 Jan 15;482(7385):414-8. doi: 10.1038/nature10744. Nature. 2012. PMID: 22246324 Free PMC article. - Structural basis for recognition of autophagic receptor NDP52 by the sugar receptor galectin-8.
Kim BW, Hong SB, Kim JH, Kwon DH, Song HK. Kim BW, et al. Nat Commun. 2013;4:1613. doi: 10.1038/ncomms2606. Nat Commun. 2013. PMID: 23511477 - LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy.
von Muhlinen N, Akutsu M, Ravenhill BJ, Foeglein Á, Bloor S, Rutherford TJ, Freund SM, Komander D, Randow F. von Muhlinen N, et al. Mol Cell. 2012 Nov 9;48(3):329-42. doi: 10.1016/j.molcel.2012.08.024. Epub 2012 Sep 27. Mol Cell. 2012. PMID: 23022382 Free PMC article. - A Structural View of Xenophagy, a Battle between Host and Microbes.
Kwon DH, Song HK. Kwon DH, et al. Mol Cells. 2018 Jan 31;41(1):27-34. doi: 10.14348/molcells.2018.2274. Epub 2018 Jan 23. Mol Cells. 2018. PMID: 29370690 Free PMC article. Review. - Dual function of CALCOCO2/NDP52 during xenophagy.
Verlhac P, Viret C, Faure M. Verlhac P, et al. Autophagy. 2015;11(6):965-6. doi: 10.1080/15548627.2015.1046672. Autophagy. 2015. PMID: 25998689 Free PMC article. Review.
Cited by
- Critical role of bacterial isochorismatase in the autophagic process induced by Acinetobacter baumannii in mammalian cells.
Wang Y, Zhang K, Shi X, Wang C, Wang F, Fan J, Shen F, Xu J, Bao W, Liu M, Yu L. Wang Y, et al. FASEB J. 2016 Oct;30(10):3563-3577. doi: 10.1096/fj.201500019R. Epub 2016 Jul 18. FASEB J. 2016. PMID: 27432399 Free PMC article. - Galectin 13 (PP13) Facilitates Remodeling and Structural Stabilization of Maternal Vessels during Pregnancy.
Sammar M, Drobnjak T, Mandala M, Gizurarson S, Huppertz B, Meiri H. Sammar M, et al. Int J Mol Sci. 2019 Jun 29;20(13):3192. doi: 10.3390/ijms20133192. Int J Mol Sci. 2019. PMID: 31261864 Free PMC article. Review. - Structural characterisation of human galectin-4 N-terminal carbohydrate recognition domain in complex with glycerol, lactose, 3'-sulfo-lactose, and 2'-fucosyllactose.
Bum-Erdene K, Leffler H, Nilsson UJ, Blanchard H. Bum-Erdene K, et al. Sci Rep. 2016 Feb 1;6:20289. doi: 10.1038/srep20289. Sci Rep. 2016. PMID: 26828567 Free PMC article. - Dining in: intracellular bacterial pathogen interplay with autophagy.
Winchell CG, Steele S, Kawula T, Voth DE. Winchell CG, et al. Curr Opin Microbiol. 2016 Feb;29:9-14. doi: 10.1016/j.mib.2015.09.004. Epub 2015 Oct 21. Curr Opin Microbiol. 2016. PMID: 26462048 Free PMC article. Review. - Recruitment of TBK1 to cytosol-invading Salmonella induces WIPI2-dependent antibacterial autophagy.
Thurston TL, Boyle KB, Allen M, Ravenhill BJ, Karpiyevich M, Bloor S, Kaul A, Noad J, Foeglein A, Matthews SA, Komander D, Bycroft M, Randow F. Thurston TL, et al. EMBO J. 2016 Aug 15;35(16):1779-92. doi: 10.15252/embj.201694491. Epub 2016 Jul 1. EMBO J. 2016. PMID: 27370208 Free PMC article.
References
- Mizushima N, Yoshimori T, Ohsumi Y. The role of atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. - PubMed
- Thurston TLM, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous