Endoplasmic reticulum stress sensing in the unfolded protein response - PubMed (original) (raw)
Review
Endoplasmic reticulum stress sensing in the unfolded protein response
Brooke M Gardner et al. Cold Spring Harb Perspect Biol. 2013.
Abstract
Secretory and transmembrane proteins enter the endoplasmic reticulum (ER) as unfolded proteins and exit as either folded proteins in transit to their target organelles or as misfolded proteins targeted for degradation. The unfolded protein response (UPR) maintains the protein-folding homeostasis within the ER, ensuring that the protein-folding capacity of the ER meets the load of client proteins. Activation of the UPR depends on three ER stress sensor proteins, Ire1, PERK, and ATF6. Although the consequences of activation are well understood, how these sensors detect ER stress remains unclear. Recent evidence suggests that yeast Ire1 directly binds to unfolded proteins, which induces its oligomerization and activation. BiP dissociation from Ire1 regulates this oligomeric equilibrium, ultimately modulating Ire1's sensitivity and duration of activation. The mechanistic principles of ER stress sensing are the focus of this review.
Figures
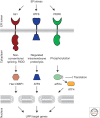
Figure 1.
In metazoans, three parallel signaling pathways comprise the UPR. ER-resident transmembrane sensor proteins, Ire1, PERK, and ATF6, activate signaling in each UPR branch. Upon activation, each sensor elicits unique downstream responses. Ire1’s cytosolic RNase domain cleaves an intron out of an mRNA, leading to the production of a potent transcriptional activator that induces genes to increase the folding capacity of the ER (XBP1 in metazoans, Hac1 in budding yeast). The active RNase also cleaves ER-localized messages, leading to their degradation, to reduce the load of unfolded proteins entering the ER. Active ATF6 translocates from the ER to the Golgi where it is proteolytically processed to release its amino-terminal transcriptional activator domain that induces genes to increase the folding capacity in the ER. PERK’s cytosolic kinase domain phosphorylates the translation initiation factor eIF2α, thereby inhibiting global translation and reducing the load of newly synthesized proteins entering the ER. Although generally inhibiting translation, eIF2α phosphorylation allows messages with inhibitory leader sequences to be preferentially translated. One of these messages encodes ATF4, a transcriptional activator that induces a cascade that ultimately produces proapoptotic factors.
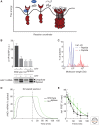
Figure 2.
Unfolded proteins are the switch that activates Ire1, while BiP binding regulates the sensitivity and duration of the response. (A) Ire1 is in equilibrium between monomeric and oligomeric states. Binding to BiP stabilizes the monomeric state by providing a sink to buffer the amount of free Ire1. Binding to unfolded proteins stabilizes the oligomeric state by overcoming the activation barrier for Ire1 oligomerization. (B) BiP dissociation from Ire1 is not sufficient to activate Ire1 (panel B is from Pincus et al. 2010; reprinted, with permission, from the authors). (Upper panel) Quantification of coimmunoprecipitation experiments showing that BiP dissociates from wild-type Ire1 in the presence of ER stress. A mutant of Ire1 lacking the ER juxtamembrane segment (Ire1bipless) binds to BiP in the absence of stress only to the degree that wild-type Ire1 binds to BiP in the presence of stress. In ER stress conditions, Ire1bipless shows no change in its association with BiP. (Lower panel) Northern blot measuring splicing of HAC1 mRNA showing that, despite its lack of ER-stress-dependent BiP dissociation, the RNase of Ire1bipless is not constitutively active and remains ER-stress inducible. (C) Peptide binding triggers Ire1-LD oligomerization in vitro (panel C is from Gardner and Walter 2011; reprinted, with permission, from the authors). Velocity sedimentation analysis of recombinant Ire1-LD in the presence and absence of a peptide proxy for an unfolded protein shows that Ire1-LD redistributes into large oligomeric assemblies in the presence of peptide. (D) Mathematical model simulation predicts that Ire1bipless would display delayed deactivation kinetics compared to wild-type Ire1 once ER stress is removed (panel D is from Pincus et al. 2010; reprinted, with permission, from the author). (E) Experimental verification that Ire1bipless deactivates less efficiently than wild-type Ire1 once ER stress is removed (panel E is from Pincus et al. 2010; reprinted, with permission, from the authors). FRET measurements between Ire1 molecules reveals that deoligomerization is impaired in the Ire1bipless mutant.
Figure 3.
Crystal structure of yeast and human Ire1-LD indicate how Ire1 oligomerizes. (A) The S. cerevisiae Ire1-LD dimer formed at Interface 1, creating a putative ligand-binding groove (PDB: 2BE1). Each monomer is colored red to blue from amino to carboxyl terminus. Compare to Figure 4A for a surface representation. (B) The H. sapiens Ire1-LD dimer formed at Interface 1 has similar architecture to yeast Ire1-LD, though the α-helical walls narrow the putative ligand binding groove (PDB: 2HZ6). Each monomer is colored red to blue from amino to carboxyl terminus. Compare to Figure 4B for a surface representation. (C) Two groove-forming dimers of yeast Ire1-LD oligomerize through Interface 2. An α-helical turn region (arrowhead) interacts with a β-sheet (arrow) to mediate this interaction. Residues that have been mutated and shown to impair UPR activation are colored red. (D) Two groove-forming dimers of human Ire1-LD interact through a novel Interface 2. The interaction between the β-sheet (arrow) and the α-helical turn region, unstructured here (arrowhead), that formed Interface 2 in the yeast Ire1-LD is now interrupted by the long helix αB. Figures were made using PyMOL (PyMOL Molecular Graphics System).
Figure 4.
Crystal structures of Ire1-LD reveal a putative ligand-binding groove similar to those of MHC and DnaK. Surface models of the structures of (A) yeast Ire1-LD, (B) human Ire1-LD, (C) MHCII peptide binding domain crystallized with a peptide ligand (magenta; PDB: 1DLH), and (D) DnaK substrate binding domain crystallized with a peptide ligand (magenta; PDB: 1DKX). All structures are colored white to blue by depth looking down on the proposed or proven ligand-binding groove. Capped side views show a cross section of the ligand-binding groove to illustrate the pockets available to bind anchor residues.
Comment in
- Endoplasmic reticulum stress and fungal pathogenesis converge.
Askew DS. Askew DS. Virulence. 2014 Feb 15;5(2):331-3. doi: 10.4161/viru.28051. Epub 2014 Feb 6. Virulence. 2014. PMID: 24504109 Free PMC article. No abstract available.
Similar articles
- Endoplasmic reticulum stress and fungal pathogenesis converge.
Askew DS. Askew DS. Virulence. 2014 Feb 15;5(2):331-3. doi: 10.4161/viru.28051. Epub 2014 Feb 6. Virulence. 2014. PMID: 24504109 Free PMC article. No abstract available. - Molecular signal networks and regulating mechanisms of the unfolded protein response.
Gong J, Wang XZ, Wang T, Chen JJ, Xie XY, Hu H, Yu F, Liu HL, Jiang XY, Fan HD. Gong J, et al. J Zhejiang Univ Sci B. 2017 Jan.;18(1):1-14. doi: 10.1631/jzus.B1600043. J Zhejiang Univ Sci B. 2017. PMID: 28070992 Free PMC article. Review. - Imaging of single cell responses to ER stress indicates that the relative dynamics of IRE1/XBP1 and PERK/ATF4 signalling rather than a switch between signalling branches determine cell survival.
Walter F, Schmid J, Düssmann H, Concannon CG, Prehn JH. Walter F, et al. Cell Death Differ. 2015 Sep;22(9):1502-16. doi: 10.1038/cdd.2014.241. Epub 2015 Jan 30. Cell Death Differ. 2015. PMID: 25633195 Free PMC article. - UPR proteins IRE1 and PERK switch BiP from chaperone to ER stress sensor.
Kopp MC, Larburu N, Durairaj V, Adams CJ, Ali MMU. Kopp MC, et al. Nat Struct Mol Biol. 2019 Nov;26(11):1053-1062. doi: 10.1038/s41594-019-0324-9. Epub 2019 Nov 6. Nat Struct Mol Biol. 2019. PMID: 31695187 Free PMC article. - The Unfolded Protein Response and Cell Fate Control.
Hetz C, Papa FR. Hetz C, et al. Mol Cell. 2018 Jan 18;69(2):169-181. doi: 10.1016/j.molcel.2017.06.017. Epub 2017 Nov 5. Mol Cell. 2018. PMID: 29107536 Review.
Cited by
- Berberine Attenuates Cerebral Ischemia-Reperfusion Injury Induced Neuronal Apoptosis by Down-Regulating the CNPY2 Signaling Pathway.
Zhao L, Li H, Gao Q, Xu J, Zhu Y, Zhai M, Zhang P, Shen N, Di Y, Wang J, Chen T, Huang M, Sun J, Liu C. Zhao L, et al. Front Pharmacol. 2021 Apr 28;12:609693. doi: 10.3389/fphar.2021.609693. eCollection 2021. Front Pharmacol. 2021. PMID: 33995012 Free PMC article. - Saikosaponin D Inducing Apoptosis and Autophagy through the Activation of Endoplasmic Reticulum Stress in Glioblastoma.
Liu G, Guan Y, Liu Y, Wang Y, Zhang J, Liu Y, Liu X. Liu G, et al. Biomed Res Int. 2022 Nov 24;2022:5489553. doi: 10.1155/2022/5489553. eCollection 2022. Biomed Res Int. 2022. PMID: 36467888 Free PMC article. - iRhom pseudoproteases regulate ER stress-induced cell death through IP3 receptors and BCL-2.
Dulloo I, Atakpa-Adaji P, Yeh YC, Levet C, Muliyil S, Lu F, Taylor CW, Freeman M. Dulloo I, et al. Nat Commun. 2022 Mar 10;13(1):1257. doi: 10.1038/s41467-022-28930-4. Nat Commun. 2022. PMID: 35273168 Free PMC article. - Naringin Prevents Cognitive Dysfunction in Aging Rats by Inhibiting Toll-Like Receptor 4 (TLR4)/NF-_κ_B Pathway and Endoplasmic Reticulum Stress.
Dai XJ, Jia Y, Cao R, Zhou MN. Dai XJ, et al. Evid Based Complement Alternat Med. 2023 Feb 21;2023:2919811. doi: 10.1155/2023/2919811. eCollection 2023. Evid Based Complement Alternat Med. 2023. PMID: 36865741 Free PMC article. - Astragaloside attenuates the progression of prostate cancer cells through endoplasmic reticulum stress pathways.
Tan B, Jia R, Wang G, Yang J. Tan B, et al. Oncol Lett. 2018 Sep;16(3):3901-3906. doi: 10.3892/ol.2018.9071. Epub 2018 Jul 4. Oncol Lett. 2018. PMID: 30128005 Free PMC article.
References
- Abravaya K, Myers MP, Murphy SP, Morimoto RI 1992. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev 6: 1153–1164 - PubMed
- Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K 2008. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct 33: 75–89 - PubMed
- Adams CM, Reitz J, De Brabander JK, Feramisco JD, Li L, Brown MS, Goldstein JL 2004. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and insigs. J Biol Chem 279: 52772–52780 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 GM103311/GM/NIGMS NIH HHS/United States
- P41 RR001081/RR/NCRR NIH HHS/United States
- 9P41GM103311/GM/NIGMS NIH HHS/United States
- 2P41RR001081/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases