Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state - PubMed (original) (raw)
Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state
Adrien Le Thomas et al. Genes Dev. 2013.
Abstract
In the metazoan germline, piwi proteins and associated piwi-interacting RNAs (piRNAs) provide a defense system against the expression of transposable elements. In the cytoplasm, piRNA sequences guide piwi complexes to destroy complementary transposon transcripts by endonucleolytic cleavage. However, some piwi family members are nuclear, raising the possibility of alternative pathways for piRNA-mediated regulation of gene expression. We found that Drosophila Piwi is recruited to chromatin, colocalizing with RNA polymerase II (Pol II) on polytene chromosomes. Knockdown of Piwi in the germline increases expression of transposable elements that are targeted by piRNAs, whereas protein-coding genes remain largely unaffected. Derepression of transposons upon Piwi depletion correlates with increased occupancy of Pol II on their promoters. Expression of piRNAs that target a reporter construct results in a decrease in Pol II occupancy and an increase in repressive H3K9me3 marks and heterochromatin protein 1 (HP1) on the reporter locus. Our results indicate that Piwi identifies targets complementary to the associated piRNA and induces transcriptional repression by establishing a repressive chromatin state when correct targets are found.
Figures
Figure 1.
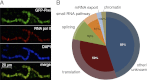
Piwi associates with chromatin and nuclear transcripts. (A) Polytene chromosomes from Drosophila nurse cells expressing GFP-Piwi on the otu[7]/otu[11] background. Piwi pattern on chromosomes correlates with Pol II staining. (B) Mass spectrometry analysis of Piwi interaction partners. Piwi complexes were precipitated in the presence and absence of RNase A. The outer circle represents classification of Piwi-associated proteins based on GO term analysis. The inner pies represent the fraction of each group whose association with Piwi depends on RNA (percentage indicated). Note that chromatin, splice, and mRNA export factors are virtually absent after RNase A treatment.
Figure 2.
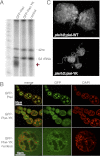
Piwi function, but not its nuclear localization, requires piRNA association. (A) The Piwi-YK mutant does not associate with piRNA. Immunoprecipitation of Piwi–piRNA complexes was performed with GFP antibody on ovaries from GFP-Piwi and GFP-Piwi-YK transgenic flies and a control strain. Small RNAs were isolated, 5′-labeled, and resolved on a denaturing gel. The same amount of 42-nucleotide RNA oligonucleotides was spiked into all samples prior to RNA isolation to control for loss of RNA during isolation and labeling. piRNAs (red arrow) are absent in the Piwi-YK complex. (B) GFP-Piwi-YK is present in the nuclei of nurse cells and colocalizes with chromatin (DAPI-stained areas). (C) The Piwi-YK mutant does not rescue the morphological changes caused by the piwi-null mutation. Dark-field images of ovaries where either the wild-type piwi or the piwi-YK transgene has been backcrossed onto the piwi-null background.
Figure 3.
Piwi transcriptionally represses TEs. (A) Piwi knockdown is efficient and specific to ovarian germ cells as indicated by GFP-Piwi localization. GFP-Piwi; Nanos-Gal4-VP16 flies were crossed to control shRNA (shWhite) or shPiwi lines. Piwi is specifically depleted in germ cells and not in follicular cells, consistent with expression of the Nanos-Gal4-VP16 driver. (B) Piwi expression as measured by RNA-seq in the Piwi knockdown and control lines. Note that Piwi expression is unaffected in follicular cells, leading to relatively weak apparent knockdown in RNA-seq libraries from whole ovaries. (C) Effect of Piwi knockdown on the expression of TEs. Two biological replicate RNA-seq experiments were carried out, and differential expression was assessed using DESeq. Transposons that show significant change (P < 0.05) are indicated by dark-red circles. Out of 217 individual RepeatMasker-annotated TEs, 15 show a significant increase in expression upon Piwi knockdown. (D) The change in the levels of TE transcripts and Pol II occupancy on their promoters upon Piwi knockdown. Twenty up-regulated and 10 down-regulated transposons with the most significant changes in expression level are shown. Note the low statistical significance for down-regulated transposons. For a complete list of transposons, see Supplemental Figure S2. (E) Pol II signal over the Het-A retrotransposon in control flies (shWhite; red) and upon Piwi knockdown (shPiwi; blue). (F) Increased abundance of transposon transcripts upon Piwi depletion correlates with increased Pol II occupancy over their promoters (_r_2 = 0.21). Note that the majority of elements do not show significant change in either RNA abundance or Pol II occupancy.
Figure 4.
Piwi-induced transcriptional repression correlates with establishment of a repressive chromatin state. (A) Overlap between genomic regions of H3K9me3 depletion upon Piwi knockdown and TEs. Two replicates of H3K9me3 ChIP-seq experiments were carried out on control and Piwi-depleted ovaries, and enriched regions were identified using DESeq (see the Materials and Methods for details). A total of 705 regions show significant (P < 0.05) decrease in H3K9me3 occupancy upon Piwi knockdown, while only 30 regions showed a similarly significant increase. Out of the 705 regions that show a decrease in H3K9me3 marks upon Piwi knockdown, 91% (646) overlap with TE sequences compared with the 33% expected from random genome sampling. (B) Genomic positions of H3K9me3-depleted regions upon Piwi depletion (outer circle) and RepeatMasker-annotated transposons (inner circle). Note that almost all regions are localized in heterochromatic and repeat-rich portions of the genome (Het, chrU, and chrUExtra chromosomes).
Figure 5.
Piwi does not directly repress protein-coding genes. (A) Effect of Piwi knockdown on the expression of genes. Two replicate RNA-seq experiments were carried out, and differential expression was assessed using DESeq. Genes that show significant change (P < 0.05) are indicated by black circles. The vast majority of genes does not change significantly upon germline Piwi knockdown (shPiwi) compared with control (shWhite). (B) H3K9me3 mark density does not change over genes that show a significant change in expression upon Piwi knockdown (see Fig. 3C). Up-regulated and down-regulated genes are plotted separately. Signal indicated is after background subtraction. (C) Functional analysis of up-regulated genes by the Database for Annotation, Visualization, and Integrated Discovery (DAVID) reveals activation of the protein degradation and DNA damage response pathways. Percentages of all up-regulated genes are indicated.
Figure 6.
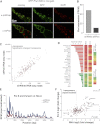
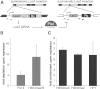
piRNA-dependent targeting of Piwi to a reporter locus leads to establishment of a repressive chromatin state and transcriptional silencing. (A) The mechanism of _trans_-silencing mediated by artificial piRNA and a schematic representation of the repressor and reporter lacZ constructs. The repressor construct is inserted in a subtelomeric piRNA cluster, leading to generation of piRNA from its sequence. Primers mapping to both constructs used for the Pol II and H3K4me2/3 ChIP-quantitative PCR (qPCR) are shown by light-gray arrows; primers specific to the reporter locus used for the H3K9me3, H3K9me2, and HP1 ChIP-qPCR are indicated by dark-gray arrows. (B) piRNAs induce transcriptional repression of the lacZ reporter. Pol II and H3K4me2/3 signals decreased on the lacZ promoter in the presence of artificial piRNAs as measured by ChIP-qPCR. Shown is the fold depletion of signal in flies that carry both repressor and reporter constructs compared with control flies that have only the reporter construct. The signal was normalized to RP49. (C) piRNAs induce an increase in H3K9me3 and H3K9me2 marks and HP1 binding as measured by ChIP-qPCR. Shown is the fold increase of corresponding ChIP signals downstream from the lacZ reporter in flies that carry both repressor and reporter constructs compared with control flies that have only reporter construct. The signal was normalized to RP49.
Comment in
- Small RNA-directed silencing: the fly finds its inner fission yeast?
Ge DT, Zamore PD. Ge DT, et al. Curr Biol. 2013 Apr 22;23(8):R318-20. doi: 10.1016/j.cub.2013.03.033. Curr Biol. 2013. PMID: 23618667
Similar articles
- Impact of nuclear Piwi elimination on chromatin state in Drosophila melanogaster ovaries.
Klenov MS, Lavrov SA, Korbut AP, Stolyarenko AD, Yakushev EY, Reuter M, Pillai RS, Gvozdev VA. Klenov MS, et al. Nucleic Acids Res. 2014 Jun;42(10):6208-18. doi: 10.1093/nar/gku268. Epub 2014 Apr 29. Nucleic Acids Res. 2014. PMID: 24782529 Free PMC article. - Piwi-piRNA complexes induce stepwise changes in nuclear architecture at target loci.
Iwasaki YW, Sriswasdi S, Kinugasa Y, Adachi J, Horikoshi Y, Shibuya A, Iwasaki W, Tashiro S, Tomonaga T, Siomi H. Iwasaki YW, et al. EMBO J. 2021 Sep 15;40(18):e108345. doi: 10.15252/embj.2021108345. Epub 2021 Aug 2. EMBO J. 2021. PMID: 34337769 Free PMC article. - piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire.
Senti KA, Jurczak D, Sachidanandam R, Brennecke J. Senti KA, et al. Genes Dev. 2015 Aug 15;29(16):1747-62. doi: 10.1101/gad.267252.115. Genes Dev. 2015. PMID: 26302790 Free PMC article. - To export, or not to export: how nuclear export factor variants resolve Piwi's dilemma.
Wang S, Lu X, Qiu D, Yu Y. Wang S, et al. Biochem Soc Trans. 2021 Nov 1;49(5):2073-2079. doi: 10.1042/BST20201171. Biochem Soc Trans. 2021. PMID: 34643228 Review. - Two distinct transcriptional controls triggered by nuclear Piwi-piRISCs in the Drosophila piRNA pathway.
Sato K, Siomi MC. Sato K, et al. Curr Opin Struct Biol. 2018 Dec;53:69-76. doi: 10.1016/j.sbi.2018.06.005. Epub 2018 Jul 7. Curr Opin Struct Biol. 2018. PMID: 29990672 Review.
Cited by
- Epigenetic regulation of germ cells-remember or forget?
Feng L, Chen X. Feng L, et al. Curr Opin Genet Dev. 2015 Apr;31:20-7. doi: 10.1016/j.gde.2015.04.003. Epub 2015 May 1. Curr Opin Genet Dev. 2015. PMID: 25930104 Free PMC article. Review. - Paramutation and related phenomena in diverse species.
Hollick JB. Hollick JB. Nat Rev Genet. 2017 Jan;18(1):5-23. doi: 10.1038/nrg.2016.115. Epub 2016 Oct 17. Nat Rev Genet. 2017. PMID: 27748375 Review. - MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle Cells.
Mugat B, Akkouche A, Serrano V, Armenise C, Li B, Brun C, Fulga TA, Van Vactor D, Pélisson A, Chambeyron S. Mugat B, et al. PLoS Genet. 2015 May 19;11(5):e1005194. doi: 10.1371/journal.pgen.1005194. eCollection 2015 May. PLoS Genet. 2015. PMID: 25993106 Free PMC article. - The piRNA pathway is developmentally regulated during spermatogenesis in Drosophila.
Quénerch'du E, Anand A, Kai T. Quénerch'du E, et al. RNA. 2016 Jul;22(7):1044-54. doi: 10.1261/rna.055996.116. Epub 2016 May 20. RNA. 2016. PMID: 27208314 Free PMC article. - Looking at the Pretty "Phase" of Membraneless Organelles: A View From Drosophila Glia.
Arkov AL. Arkov AL. Front Cell Dev Biol. 2022 Feb 7;10:801953. doi: 10.3389/fcell.2022.801953. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35198559 Free PMC article. Review.
References
- Agger K, Cloos P, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini A, Helin K 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734 - PubMed
- Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau J-C et al. 2012. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol 19: 998–1004 - PubMed
- Aravin A, Hannon G, Brennecke J 2007. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM007616/GM/NIGMS NIH HHS/United States
- R00 HD057233/HD/NICHD NIH HHS/United States
- DP2OD007371A/OD/NIH HHS/United States
- DP2 OD007371/OD/NIH HHS/United States
- NIH/NRSA 5T32 GM07616/GM/NIGMS NIH HHS/United States
- R01 GM097363/GM/NIGMS NIH HHS/United States
- U54 HG004576/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials