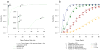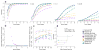Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples - PubMed (original) (raw)
Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples
Kristian Cibulskis et al. Nat Biotechnol. 2013 Mar.
Abstract
Detection of somatic point substitutions is a key step in characterizing the cancer genome. However, existing methods typically miss low-allelic-fraction mutations that occur in only a subset of the sequenced cells owing to either tumor heterogeneity or contamination by normal cells. Here we present MuTect, a method that applies a Bayesian classifier to detect somatic mutations with very low allele fractions, requiring only a few supporting reads, followed by carefully tuned filters that ensure high specificity. We also describe benchmarking approaches that use real, rather than simulated, sequencing data to evaluate the sensitivity and specificity as a function of sequencing depth, base quality and allelic fraction. Compared with other methods, MuTect has higher sensitivity with similar specificity, especially for mutations with allelic fractions as low as 0.1 and below, making MuTect particularly useful for studying cancer subclones and their evolution in standard exome and genome sequencing data.
Figures
Figure 1
Overview of somatic point mutation detection using MuTect. MuTect takes as input tumor (T) and normal (N) next generation sequencing data and, after removing low quality reads (Supplementary Methods), determines if there is evidence for a variant beyond the expected random sequencing errors. Candidate variant sites are then passed through six filters to remove artifacts (Table 1). Next, a Panel of Normals is used to screen out remaining false positives caused by rare error modes only detectable in additional samples. Finally, the somatic or germline status of passing variants is determined using the matched normal.
Figure 2
Sensitivity as a function of sequencing depth and allelic fraction. (a) Sensitivity and specificity of MuTect for mutations with an allele fraction of 0.2, tumor depth of 30x and normal depth of 30x using various values of the LOD threshold (θT) (0.1 ≤ θT ≤ 100). Results using a model of independent sequencing errors with uniform Q35 base quality scores and accurate read placement (solid grey) are shown as well as results from the virtual tumor approach for the standard (STD, dashed green) and high-confidence (HC, solid green) configurations. A typical setting of θT = 6.3 is marked with black circles. (b) Sensitivity as a function of tumor sequencing depth and allele fraction (indicated by color) using θT = 6.3. The calculated sensitivity using a model of independent sequencing errors and accurate read placement with uniform Q35 base quality scores (solid lines) are shown as well as results from the virtual tumor approach (circles) and the downsampling of validated colorectal mutations (diamonds). Error bars represent 95% CIs.
Figure 3
Specificity of variant detection and variant classification using virtual tumor approach. (a) Somatic miscall error rate for true reference sites as a function of tumor sequencing depth for the STD (red), HC (blue) and HC+PON (green) configurations of MuTect. Error bars represent 95% CIs. (b) Distribution of allele fraction for all miscalls as a function of tumor sequencing depth. (c) Fraction of events rejected by each filter; hashed regions indicate events rejected exclusively by each filter. (d) Somatic miscall error rate for true germline SNP sites by sequencing depth in the normal when the site is known to be variant in the population (blue) and novel (red). Error bars represent 95% CIs. (e,f) Mean power as a function of sequencing depth in the normal to have classified these events as germline or somatic at novel germline sites (e) and known germline variant sites (f).
Figure 4
Benchmarking mutation detection methods. (a) Comparison of sensitivity as a function of tumor sequencing depth and mutation allele fraction for different mutation detection methods and configurations. (b) Comparison of somatic miscall error rate for true germline sites as a function of sequencing depth in the normal. (c) Comparison of somatic miscall error rate for true reference sites as a function of tumor sequencing depth. (d) Sensitivity as a function of specificity for mutations with an allele fraction of 0.1, tumor depth of 30x and normal depth of 30x between different methods and configurations. Black dotted lines indicate change in sensitivity and specificity between STD and HC configurations for a method. Grey solid lines are the MuTect results of virtual tumor approach from Supplementary Figure 3. (a–c) Error bars represent 95% CIs.
Comment in
- Bioinformatics: Tumors have their differences.
Nawy T. Nawy T. Nat Methods. 2013 Apr;10(4):282-3. doi: 10.1038/nmeth.2426. Nat Methods. 2013. PMID: 23653923 No abstract available.
Similar articles
- Optimized pipeline of MuTect and GATK tools to improve the detection of somatic single nucleotide polymorphisms in whole-exome sequencing data.
do Valle ÍF, Giampieri E, Simonetti G, Padella A, Manfrini M, Ferrari A, Papayannidis C, Zironi I, Garonzi M, Bernardi S, Delledonne M, Martinelli G, Remondini D, Castellani G. do Valle ÍF, et al. BMC Bioinformatics. 2016 Nov 8;17(Suppl 12):341. doi: 10.1186/s12859-016-1190-7. BMC Bioinformatics. 2016. PMID: 28185561 Free PMC article. - SiNVICT: ultra-sensitive detection of single nucleotide variants and indels in circulating tumour DNA.
Kockan C, Hach F, Sarrafi I, Bell RH, McConeghy B, Beja K, Haegert A, Wyatt AW, Volik SV, Chi KN, Collins CC, Sahinalp SC. Kockan C, et al. Bioinformatics. 2017 Jan 1;33(1):26-34. doi: 10.1093/bioinformatics/btw536. Epub 2016 Aug 16. Bioinformatics. 2017. PMID: 27531099 - Integrated approach to generate artificial samples with low tumor fraction for somatic variant calling benchmarking.
Sergi A, Beltrame L, Marchini S, Masseroli M. Sergi A, et al. BMC Bioinformatics. 2024 May 8;25(1):180. doi: 10.1186/s12859-024-05793-8. BMC Bioinformatics. 2024. PMID: 38720249 Free PMC article. - Targeted deep resequencing of the human cancer genome using next-generation technologies.
Myllykangas S, Ji HP. Myllykangas S, et al. Biotechnol Genet Eng Rev. 2010;27:135-58. doi: 10.1080/02648725.2010.10648148. Biotechnol Genet Eng Rev. 2010. PMID: 21415896 Free PMC article. Review. - Computational approaches for discovery of mutational signatures in cancer.
Baez-Ortega A, Gori K. Baez-Ortega A, et al. Brief Bioinform. 2019 Jan 18;20(1):77-88. doi: 10.1093/bib/bbx082. Brief Bioinform. 2019. PMID: 28968631 Free PMC article. Review.
Cited by
- Genomic and transcriptomic analyses identify distinctive features of triple-negative inflammatory breast cancer.
Wang X, Zhao L, Song X, Wu X, Krishnamurthy S, Semba T, Shao S, Knafl M, Coffer LW 2nd, Alexander A, Vines A, Bopparaju S, Woodward WA, Chu R, Zhang J, Yam C, Loo LWM, Nasrazadani A, Huong LP, Woodman SE, Futreal A; Rare Tumor Initiative Team; Tripathy D, Ueno NT. Wang X, et al. NPJ Precis Oncol. 2024 Nov 18;8(1):265. doi: 10.1038/s41698-024-00729-0. NPJ Precis Oncol. 2024. PMID: 39558017 Free PMC article. - Whole-exome sequencing reveals genomic landscape of intrahepatic cholangiocarcinoma and identifies SAV1 as a potential driver.
Zhou ZJ, Ye YH, Hu ZQ, Hou YR, Liu KX, Sun RQ, Wang PC, Luo CB, Li J, Zou JX, Zhou J, Fan J, Song CL, Zhou SL. Zhou ZJ, et al. Nat Commun. 2024 Nov 17;15(1):9960. doi: 10.1038/s41467-024-54387-8. Nat Commun. 2024. PMID: 39551842 Free PMC article. - Molecular profiling reveals novel therapeutic targets and clonal evolution in ovarian clear cell carcinoma.
Chao A, Huang CY, Yu W, Lin CY, Lin H, Chao AS, Lin CT, Chou HH, Huang KG, Huang HJ, Chang TC, Rozen SG, Wu RC, Lai CH. Chao A, et al. BMC Cancer. 2024 Nov 14;24(1):1403. doi: 10.1186/s12885-024-13125-5. BMC Cancer. 2024. PMID: 39543535 Free PMC article. - Genomic profiling of circulating tumor DNA for childhood cancers.
Lei S, Jia S, Takalkar S, Chang TC, Ma X, Szlachta K, Xu K, Cheng Z, Hui Y, Koo SC, Mead PE, Gao Q, Kumar P, Bailey CP, Sunny J, Pappo AS, Federico SM, Robinson GW, Gajjar A, Rubnitz JE, Jeha S, Pui CH, Inaba H, Wu G, Klco JM, Tatevossian RG, Mullighan CG. Lei S, et al. Leukemia. 2024 Nov 10. doi: 10.1038/s41375-024-02461-x. Online ahead of print. Leukemia. 2024. PMID: 39523434 - Oncolytic immunotherapy with nivolumab in muscle-invasive bladder cancer: a phase 1b trial.
Li R, Villa NY, Yu X, Johnson JO, Borjas G, Dhillon J, Moran-Segura CM, Kim Y, Francis N, Dorman D, Powers JJ, Sexton WJ, Spiess PE, Poch MA, Zemp L, Gilbert SM, Zhang J, Pow-Sang JM, Anderson ARA, Li T, Wang X, Grass GD, Burke JM, Dinney CPN, Rodriguez PC, Jain RK, Mulé JJ, Conejo-Garcia JR. Li R, et al. Nat Med. 2024 Nov 9. doi: 10.1038/s41591-024-03324-9. Online ahead of print. Nat Med. 2024. PMID: 39521884
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources