Disclosure of a structural milieu for the proximity ligation reveals the elusive nature of an active chromatin hub - PubMed (original) (raw)
Disclosure of a structural milieu for the proximity ligation reveals the elusive nature of an active chromatin hub
Alexey A Gavrilov et al. Nucleic Acids Res. 2013.
Abstract
The current progress in the study of the spatial organization of interphase chromosomes became possible owing to the development of the chromosome conformation capture (3C) protocol. The crucial step of this protocol is the proximity ligation-preferential ligation of DNA fragments assumed to be joined within nuclei by protein bridges and solubilized as a common complex after formaldehyde cross-linking and DNA cleavage. Here, we show that a substantial, and in some cases the major, part of DNA is not solubilized from cross-linked nuclei treated with restriction endonuclease(s) and sodium dodecyl sulphate and that this treatment neither causes lysis of the nucleus nor drastically affects its internal organization. Analysis of the ligation frequencies of the mouse β-globin gene domain DNA fragments demonstrated that the previously reported 3C signals were generated predominantly, if not exclusively, in the insoluble portion of the 3C material. The proximity ligation thus occurs within the cross-linked chromatin cage in non-lysed nuclei. The finding does not compromise the 3C protocol but allows the consideration of an active chromatin hub as a folded chromatin domain or a nuclear compartment rather than a rigid complex of regulatory elements.
Figures
Figure 1.
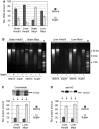
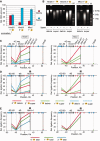
Partitioning of DNA and histones between soluble and insoluble portions of the 3C material and size distribution of DNA fragments. (A) Relative amounts of DNA in soluble (super) and insoluble (debris) portions of the 3C material, as determined by fluorometric assays (Qubit, Invitrogen). In each experiment, the total amount of DNA in two fractions is set as 100. (B) Electrophoretic separation of DNA from soluble and insoluble portions of the 3C material before and after ligation (agarose gel, ethidium bromide staining). M—DNA size marker (Fermentas, SM0331). (C and D) Partitioning of histones between the soluble and the insoluble portions of the 3C material. Proteins present in equal portions of the soluble and the insoluble 3C material were separated by PAAG and visualized by Coomassie staining (C) or by immunoblotting with antibodies against histone H3 (D). The intensity of bands was quantified using ImageJ software. In each experiment, the total amount of histones in two fractions is set as 100. The error bars represent SEM for three independent experiments.
Figure 2.
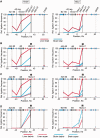
Frequencies of ligation of the fragment harboring the Hbb-b1 promoter with several selected fragments of the β-globin gene domain in soluble and insoluble portions of the 3C material. (A) Results of standard 3C analysis performed without fractionating the 3C material. (B) Results of 3C analysis performed separately on soluble (super) and insoluble (debris) fractions. (C) The same as (B) after normalization of the ligation frequencies to the amount of DNA in the samples. (D) The same as (C), soluble fraction only. On the top of each graph, a map of the domain is shown, with β-globin genes, olfactory receptor genes and DNase I hypersensitive sites shown by red arrows, blue arrows and black vertical lines, respectively. Plotted on the horizontal axis are the fragment positions. The scale is in kilobases, and according to GenBank entry NT_039433, the ‘0’ point corresponds to the start of the Hbb-y gene. The black rectangle in the background of each graph shows the anchor fragment, and the gray rectangles indicate test fragments. Plotted on the vertical axis are the ligation frequencies; the highest ligation frequency observed is set to 100 [the frequency of ligation between the anchor fragment and the upstream restriction fragment in the total 3C material from fetal liver cells (A) or in the insoluble portion of the 3C material from fetal liver cells (B and C) or the soluble portion of the 3C material from fetal brain cells (D)]. Red and blue lines show the results for liver and brain cells, respectively; solid lines show the results for the total 3C material (A) or the insoluble portion of the 3C material (B and C); dotted lines show the results for the soluble portion of the 3C material. Ligation frequencies of HindIII and MboI fragments are presented on the left and the right graphs, respectively. The error bars represent SEM for three independent experiments.
Figure 3.
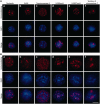
Visualization of nuclear compartments and chromatin domains in non-treated liver cells (A) and the same cells treated according to the 3C protocol up to the ligation step (B). The insoluble fraction was collected after HindIII digestion and 1.6% SDS extraction. (a–e) Immunostaining with antibodies against nucleolin (a), Sc35 (b), DNA topoisomerase II (c), H3K9me3 (d) and H3K27me3 (e). (f) Visualization of the chromosome 7 territory (FISH with a library of the chromosome 7–specific probes). In both sections of the Figure, the results of immunostaining are shown in the first row (red) and counterstaining of DNA with DAPI is shown in the second row (blue). The superimposition of the immunostaining and counterstaining of DNA is shown in the third row. Scale bar: 5 µm.
Figure 4.
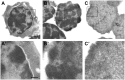
Electron microscopic analysis of the insoluble 3C material from liver cells at different steps of the 3C procedure. After formaldehyde cross-linking (A and A’), after isolation of nuclei and extraction with 0.3% SDS followed by 1.8% Triton X-100 (B and B’) and after digestion with HindIII restriction endonuclease followed by extraction with 1.6% SDS (C and C’). Panels below show the enlarged framed region of the above images. Scale bars: 1 µm (A–C) and 250 nm (A’–C’).
Figure 5.
Effect of sonication on the DNA partitioning between soluble and insoluble portions of the 3C material and on the frequencies of ligation of the β-globin gene domain fragments. (A) Relative amounts of DNA in the soluble and the insoluble portions of the sonicated 3C material. (B) Electrophoretic separation of DNA from soluble and insoluble portions of the sonicated 3C material before and after ligation. (C) Results of 3C analysis performed separately on soluble (super) and insoluble (debris) portions of the sonicated 3C material. (D) The same as (C) after normalization of the ligation frequencies to the amount of DNA in the samples. (E) Results of standard 3C analysis performed on the sonicated 3C material without fractionating into the soluble and the insoluble portions. The sonication time is indicated in seconds. The error bars represent SEM for two independent experiments. Other designations are as in Figure 1 and 2.
Figure 6.
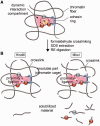
The proposed model of an active chromatin hub/compartment. (A) A schematic showing a putative dynamic interaction compartment containing two promoters (P1 and P2) controlled by a distant enhancer (enh). The enhancer is located in close spatial proximity to P1 and P2 owing to the formation of a chromatin loop stabilized by a cohesin ring (blue ring in the schematic). A complex of regulatory proteins (yellow circles) may or may not directly join the enhancer and promoter(s). After formaldehyde fixation, the mutual positions of the enhancer and the promoters became ‘frozen’ owing to the cross-links between closely located chromatin fibers. For the simplicity of presentation, these fibers are shown crossing each other. (B) After SDS extraction, the putative multicomponent protein complexes joining the enhancer and the promoter(s) are disintegrated (because of non-effective fixation of such complexes by formaldehyde), while the chromatin ‘cage’ stabilized by formaldehyde cross-links survives. Cleavage of DNA with restriction enzymes produces cohesive ends that possess a certain (limited) mobility within a chromatin cage so that the cohesive ends of different chromatin/DNA fibers located in close spatial proximity can be cross-ligated. Linking of chromatin fibers by formaldehyde is a stochastic process. In our model, the interaction compartment is stabilized by four cross-links between chromatin fibers. A failure to produce some of these cross-links will result in the separation of DNA fragments bearing the enhancer or the promoters from a chromatin cage. These fragments will be solubilized. However, in a solution, the fragments bearing the enhancer and the promoters will no longer be held in spatial proximity. Consequently, ligation of solubilized fragments will proceed without special preference and will not result in the generation of characteristic 3C signals.
Similar articles
- Actual ligation frequencies in the chromosome conformation capture procedure.
Gavrilov AA, Golov AK, Razin SV. Gavrilov AA, et al. PLoS One. 2013;8(3):e60403. doi: 10.1371/journal.pone.0060403. Epub 2013 Mar 26. PLoS One. 2013. PMID: 23555968 Free PMC article. - Mapping of the nuclear matrix-bound chromatin hubs by a new M3C experimental procedure.
Gavrilov AA, Zukher IS, Philonenko ES, Razin SV, Iarovaia OV. Gavrilov AA, et al. Nucleic Acids Res. 2010 Dec;38(22):8051-60. doi: 10.1093/nar/gkq712. Epub 2010 Aug 12. Nucleic Acids Res. 2010. PMID: 20705651 Free PMC article. - Chromosome Conformation Capture (3C) in Budding Yeast.
Belton JM, Dekker J. Belton JM, et al. Cold Spring Harb Protoc. 2015 Jun 1;2015(6):580-6. doi: 10.1101/pdb.prot085175. Cold Spring Harb Protoc. 2015. PMID: 26034304 - Close encounters of the 3C kind: long-range chromatin interactions and transcriptional regulation.
Palstra RJ. Palstra RJ. Brief Funct Genomic Proteomic. 2009 Jul;8(4):297-309. doi: 10.1093/bfgp/elp016. Epub 2009 Jun 17. Brief Funct Genomic Proteomic. 2009. PMID: 19535505 Review. - Chromosome conformation capture (from 3C to 5C) and its ChIP-based modification.
Gavrilov A, Eivazova E, Priozhkova I, Lipinski M, Razin S, Vassetzky Y. Gavrilov A, et al. Methods Mol Biol. 2009;567:171-88. doi: 10.1007/978-1-60327-414-2_12. Methods Mol Biol. 2009. PMID: 19588093 Review.
Cited by
- 3D imaging of Sox2 enhancer clusters in embryonic stem cells.
Liu Z, Legant WR, Chen BC, Li L, Grimm JB, Lavis LD, Betzig E, Tjian R. Liu Z, et al. Elife. 2014 Dec 24;3:e04236. doi: 10.7554/eLife.04236. Elife. 2014. PMID: 25537195 Free PMC article. - Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization.
Williamson I, Berlivet S, Eskeland R, Boyle S, Illingworth RS, Paquette D, Dostie J, Bickmore WA. Williamson I, et al. Genes Dev. 2014 Dec 15;28(24):2778-91. doi: 10.1101/gad.251694.114. Genes Dev. 2014. PMID: 25512564 Free PMC article. - Anthrax Susceptibility: Human Genetic Polymorphisms Modulating ANTXR2 Expression.
Zhang Z, Zhang Y, Shi M, Ye B, Shen W, Li P, Xing L, Zhang X, Hou L, Xu J, Zhao Z, Chen W. Zhang Z, et al. Toxins (Basel). 2015 Dec 22;8(1):1. doi: 10.3390/toxins8010001. Toxins (Basel). 2015. PMID: 26703731 Free PMC article. - Mapping 3D genome architecture through in situ DNase Hi-C.
Ramani V, Cusanovich DA, Hause RJ, Ma W, Qiu R, Deng X, Blau CA, Disteche CM, Noble WS, Shendure J, Duan Z. Ramani V, et al. Nat Protoc. 2016 Nov;11(11):2104-21. doi: 10.1038/nprot.2016.126. Epub 2016 Sep 29. Nat Protoc. 2016. PMID: 27685100 Free PMC article. - The missing story behind Genome Wide Association Studies: single nucleotide polymorphisms in gene deserts have a story to tell.
Schierding W, Cutfield WS, O'Sullivan JM. Schierding W, et al. Front Genet. 2014 Feb 18;5:39. doi: 10.3389/fgene.2014.00039. eCollection 2014. Front Genet. 2014. PMID: 24600475 Free PMC article. Review.
References
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. - PubMed
- Ethier SD, Miura H, Dostie J. Discovering genome regulation with 3C and 3C-related technologies. Biochim. Biophys. Acta. 2012;1819:401–410. - PubMed
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002;10:1453–1465. - PubMed
- de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–459. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources