Diverse and abundant antibiotic resistance genes in Chinese swine farms - PubMed (original) (raw)
Diverse and abundant antibiotic resistance genes in Chinese swine farms
Yong-Guan Zhu et al. Proc Natl Acad Sci U S A. 2013.
Abstract
Antibiotic resistance genes (ARGs) are emerging contaminants posing a potential worldwide human health risk. Intensive animal husbandry is believed to be a major contributor to the increased environmental burden of ARGs. Despite the volume of antibiotics used in China, little information is available regarding the corresponding ARGs associated with animal farms. We assessed type and concentrations of ARGs at three stages of manure processing to land disposal at three large-scale (10,000 animals per year) commercial swine farms in China. In-feed or therapeutic antibiotics used on these farms include all major classes of antibiotics except vancomycins. High-capacity quantitative PCR arrays detected 149 unique resistance genes among all of the farm samples, the top 63 ARGs being enriched 192-fold (median) up to 28,000-fold (maximum) compared with their respective antibiotic-free manure or soil controls. Antibiotics and heavy metals used as feed supplements were elevated in the manures, suggesting the potential for coselection of resistance traits. The potential for horizontal transfer of ARGs because of transposon-specific ARGs is implicated by the enrichment of transposases--the top six alleles being enriched 189-fold (median) up to 90,000-fold in manure--as well as the high correlation (r(2) = 0.96) between ARG and transposase abundance. In addition, abundance of ARGs correlated directly with antibiotic and metal concentrations, indicating their importance in selection of resistance genes. Diverse, abundant, and potentially mobile ARGs in farm samples suggest that unmonitored use of antibiotics and metals is causing the emergence and release of ARGs to the environment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
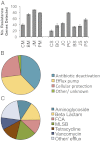
Antibiotic resistance gene detection statistics. Sample names are abbreviated with two letters representing location and sample type: first C, B, J, and P (control, Beijing, Jiaxing, and Putian, respectively) and second M, C, and S [manure, compost, and soil (with compost amendment), respectively]. Because many resistance genes were targeted with multiple primers, if multiple primer sets detected the same gene, this was only counted as detection of a single unique resistance gene. (A) Average number of unique resistance genes detected in each sample. Error bars represent SEM of four field replicates. The resistance genes detected in all samples were classified based on (B) the mechanism of resistance, and (C) the antibiotic to which they confer resistance. FCA, fluoroquinolone, quinolone, florfenicol, chloramphenicol, and amphenicol resistance genes; MLSB, Macrolide-Lincosamide-Streptogramin B resistance.
Fig. 2.
Resistance gene profile from the farm sites. Each column is labeled with the sample name (same abbreviation scheme as in Fig. 1, with numbers representing field replicates), and each row is the results from a single primer set. Values plotted are the ΔΔCT with the control soil being the reference sample for all samples. The legend denotes corresponding fold change values, which is a log scale. All primer sets (223) that showed amplification in at least one sample are shown. Columns were clustered based on Bray-Curtis diversity measures. Black boxes delineate resistance profiles: (A) enriched in all samples, including control manure (CM); (B) enriched in all farm samples, but not the CM; (C) widely enriched in most of the farm samples but not the CM; (D) genes that were enriched in the Putian compost but not the Putian manure; and (E) strongly enriched in CM and farm manures.
Fig. 3.
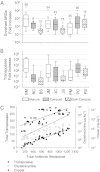
Abundance of resistance genes and transposases. In the box plots, the symbols indicate the following: box, 25th to 75th percentile; horizontal line, median; whiskers, 10th and 90th percentile; and square, maximum value. The y axis is a log scale of fold increase: farm manure compared with the control manure, and farm compost or soil compared with the control soil. (A) Only statistically enriched resistance genes are represented. The number above each site indicates the number of primer sets that yielded statistically significant results. (B) Summary of all nine primer sets used to target different transposase alleles (in B, top whisker represents the maximum value). (C) Correlation of total resistance and transposase abundances, oxytetracycline concentration, and copper concentration. Total antibiotic resistance and total transposases values are the sum of ∆∆CT values of all assays of that type in each sample. The sample identifiers below B apply to both A and B.
Fig. 4.
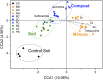
Canonical correspondence analysis (CCA) compares the abundance of detected resistance genes (symbols) and the concentration of heavy metals and antibiotics (arrows). The results showed that pig manure samples were positively correlated to the concentrations of copper, zinc, arsenic, and total tetracyclines. Environmental variables were chosen based on significance calculated from individual CCA results and variance inflation factors (VIFs) calculated during CCA. The percentage of variation explained by each axis is shown, and the relationship is significant (P = 0.005). CCA analyses were performed in R 2.13.0 with vegan package 1.17-9.
Similar articles
- Prevalence and dissemination of antibiotic resistance genes and coselection of heavy metals in Chinese dairy farms.
Zhou B, Wang C, Zhao Q, Wang Y, Huo M, Wang J, Wang S. Zhou B, et al. J Hazard Mater. 2016 Dec 15;320:10-17. doi: 10.1016/j.jhazmat.2016.08.007. Epub 2016 Aug 4. J Hazard Mater. 2016. PMID: 27505289 - Changes in antibiotic concentrations and antibiotic resistome during commercial composting of animal manures.
Xie WY, Yang XP, Li Q, Wu LH, Shen QR, Zhao FJ. Xie WY, et al. Environ Pollut. 2016 Dec;219:182-190. doi: 10.1016/j.envpol.2016.10.044. Epub 2016 Oct 27. Environ Pollut. 2016. PMID: 27814534 - Which animal type contributes the most to the emission of antibiotic resistance genes in large-scale swine farms in China?
Zhang J, Lu T, Chai Y, Sui Q, Shen P, Wei Y. Zhang J, et al. Sci Total Environ. 2019 Mar 25;658:152-159. doi: 10.1016/j.scitotenv.2018.12.175. Epub 2018 Dec 12. Sci Total Environ. 2019. PMID: 30577014 - Antibiotic resistance genes in China: occurrence, risk, and correlation among different parameters.
Zhao W, Wang B, Yu G. Zhao W, et al. Environ Sci Pollut Res Int. 2018 Aug;25(22):21467-21482. doi: 10.1007/s11356-018-2507-z. Epub 2018 Jun 12. Environ Sci Pollut Res Int. 2018. PMID: 29948704 Review. - Distribution of antibiotic resistance genes and their pathogen hosts in duck farm environments in south-east coastal China.
Liu K, Wang M, Zhang Y, Fang C, Zhang R, Fang L, Sun J, Liu Y, Liao X. Liu K, et al. Appl Microbiol Biotechnol. 2024 Dec;108(1):136. doi: 10.1007/s00253-023-12842-4. Epub 2024 Jan 15. Appl Microbiol Biotechnol. 2024. PMID: 38229327 Free PMC article. Review.
Cited by
- Fecal microbial gene transfer contributes to the high-grain diet-induced augmentation of aminoglycoside resistance in dairy cattle.
Zhang T, Mu Y, Gao Y, Tang Y, Mao S, Liu J. Zhang T, et al. mSystems. 2024 Jan 23;9(1):e0081023. doi: 10.1128/msystems.00810-23. Epub 2023 Dec 12. mSystems. 2024. PMID: 38085089 Free PMC article. - Geographic patterns and determinants of antibiotic resistomes in coastal sediments across complex ecological gradients.
Xiong S, Wang K, Yan H, Hou D, Wang Y, Li M, Zhang D. Xiong S, et al. Front Microbiol. 2022 Nov 3;13:922580. doi: 10.3389/fmicb.2022.922580. eCollection 2022. Front Microbiol. 2022. PMID: 36406438 Free PMC article. - Metal stressors consistently modulate bacterial conjugal plasmid uptake potential in a phylogenetically conserved manner.
Klümper U, Dechesne A, Riber L, Brandt KK, Gülay A, Sørensen SJ, Smets BF. Klümper U, et al. ISME J. 2017 Jan;11(1):152-165. doi: 10.1038/ismej.2016.98. Epub 2016 Aug 2. ISME J. 2017. PMID: 27482924 Free PMC article. - Characterization of the resistome in manure, soil and wastewater from dairy and beef production systems.
Noyes NR, Yang X, Linke LM, Magnuson RJ, Cook SR, Zaheer R, Yang H, Woerner DR, Geornaras I, McArt JA, Gow SP, Ruiz J, Jones KL, Boucher CA, McAllister TA, Belk KE, Morley PS. Noyes NR, et al. Sci Rep. 2016 Apr 20;6:24645. doi: 10.1038/srep24645. Sci Rep. 2016. PMID: 27095377 Free PMC article. - Occurrence and prevalence of antibiotic resistance in landfill leachate.
Wang Y, Tang W, Qiao J, Song L. Wang Y, et al. Environ Sci Pollut Res Int. 2015 Aug;22(16):12525-33. doi: 10.1007/s11356-015-4514-7. Epub 2015 Apr 24. Environ Sci Pollut Res Int. 2015. PMID: 25903180
References
- Arias CA, Murray BE. Antibiotic-resistant bugs in the 21st century—A clinical super-challenge. N Engl J Med. 2009;360(5):439–443. - PubMed
- Udwadia ZF, Amale RA, Ajbani KK, Rodrigues C. Totally drug-resistant tuberculosis in India. Clin Infect Dis. 2012;54(4):579–581. - PubMed
- Knapp CW, Dolfing J, Ehlert PAI, Graham DW. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ Sci Technol. 2010;44(2):580–587. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical