Long noncoding RNAs regulate adipogenesis - PubMed (original) (raw)
. 2013 Feb 26;110(9):3387-92.
doi: 10.1073/pnas.1222643110. Epub 2013 Feb 11.
Loyal A Goff, Cole Trapnell, Ryan Alexander, Kinyui Alice Lo, Ezgi Hacisuleyman, Martin Sauvageau, Barbara Tazon-Vega, David R Kelley, David G Hendrickson, Bingbing Yuan, Manolis Kellis, Harvey F Lodish, John L Rinn
Affiliations
- PMID: 23401553
- PMCID: PMC3587215
- DOI: 10.1073/pnas.1222643110
Long noncoding RNAs regulate adipogenesis
Lei Sun et al. Proc Natl Acad Sci U S A. 2013.
Abstract
The prevalence of obesity has led to a surge of interest in understanding the detailed mechanisms underlying adipocyte development. Many protein-coding genes, mRNAs, and microRNAs have been implicated in adipocyte development, but the global expression patterns and functional contributions of long noncoding RNA (lncRNA) during adipogenesis have not been explored. Here we profiled the transcriptome of primary brown and white adipocytes, preadipocytes, and cultured adipocytes and identified 175 lncRNAs that are specifically regulated during adipogenesis. Many lncRNAs are adipose-enriched, strongly induced during adipogenesis, and bound at their promoters by key transcription factors such as peroxisome proliferator-activated receptor γ (PPARγ) and CCAAT/enhancer-binding protein α (CEBPα). RNAi-mediated loss of function screens identified functional lncRNAs with varying impact on adipogenesis. Collectively, we have identified numerous lncRNAs that are functionally required for proper adipogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Global transcriptome profiling of lncRNAs and protein coding genes during adipogenesis via RNA-Seq. (A) This heatmap shows 1,734 coding genes and 175 lncRNAs significantly up-regulated (red) or down-regulated (blue) between preadipocytes and differentiated adipocytes. Independent hierarchical clustering of expression values for these lncRNAs and coding genes clusters differentiated and primary adipocytes together. (B) RNA-Seq read alignment of three lncRNAs as well as three adipocyte markers—AdipoQ, Fabp4, and Glut4—is displayed. RNA-Seq read alignment coverage in brown and white preadipocytes and cultured mature adipocytes is displayed above each gene. Previously identified PPARγ ChIP-Seq binding sites (10) are identified with an asterisk (*), and exons are depicted as blue bars.
Fig. 2.
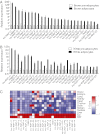
Validation of selected lncRNAs. Primary brown preadipocytes (A) and primary white preadipocytes (B) were isolated, cultured, and differentiated as described in Experimental Procedures. Total RNA was extracted from D0 and D6 cells, and qRT-PCR was performed to examine the expression of selected lncRNAs. Twenty-six lncRNAs (n = 3) were validated as significantly differentially up-regulated during both brown and white fat differentiation (P < 0.05; means ± SEM). (C) RT-PCR was performed to examine the expression of lncRNAs across 12 adult mouse tissues. Their expression profiles, presented here as differences from the row mean, are plotted as a heatmap. Red represents a higher expression and blue a lower expression. BAT, brown adipose tissue; Epi, epididymal white fat.
Fig. 3.
LncRNAs are essential regulators for adipogenesis. (A) Primary white preadipocytes were isolated, cultured, and differentiated as described in Experimental Procedures. One day before differentiation, cells were transfected with siRNAs targeting each lncRNA. At 4 d after differentiation induction, ORO staining was performed to detect the lipid accumulation. RT-PCR was performed to examine the expression of the targeted lncRNAs (A, bar plots) (n = 3; *P < 0.05; means ± SEM) and several adipogenesis marker genes (B).
Fig. 4.
Microarray analysis of adipogenesis-regulated LncRNA knockdowns. Gene expression profiles for 1,727 genes selected as significant between undifferentiated (D0) and differentiated (D4) adipose precursors but not significantly regulated between D4 and a scrambled siRNA control. Knockdowns are rank-ordered by JSD to D0, and this ordering highlights the range of effects each LncRNA has on regulating the adipogenesis transcriptome. The dashed lines divide the knockdowns into three regions: those with little to no effect on adipogenesis (similar to D4 and scramble), those with moderate effects on the adipogenesis-regulated genes (middle block), and those ncRNAs whose absence severely affects the differentiation potential of adipocyte precursors (similar to D0). Distance from D0 correlates with lipid accumulation, as determined by Oil Red staining (plates).
Similar articles
- AGPAT3 deficiency impairs adipocyte differentiation and leads to a lean phenotype in mice.
Zhou H, Fick K, Patel V, Hilton LR, Kim HW, Bagi Z, Weintraub NL, Chen W. Zhou H, et al. Am J Physiol Endocrinol Metab. 2024 Jul 1;327(1):E69-E80. doi: 10.1152/ajpendo.00012.2024. Epub 2024 May 8. Am J Physiol Endocrinol Metab. 2024. PMID: 38717361 Free PMC article. - Long Noncoding RNAs Expressed in Mouse Pituitary Development and Mature Hormone-Producing Cells.
Brinkmeier ML, George AS, Cheung LYM, Mills RE, Melamed P, Camper SA. Brinkmeier ML, et al. Endocrinology. 2024 Oct 30;165(12):bqae147. doi: 10.1210/endocr/bqae147. Endocrinology. 2024. PMID: 39487735 - New insights for precision treatment of glioblastoma from analysis of single-cell lncRNA expression.
Meng Q, Zhang Y, Li G, Li Y, Xie H, Chen X. Meng Q, et al. J Cancer Res Clin Oncol. 2021 Jul;147(7):1881-1895. doi: 10.1007/s00432-021-03584-9. Epub 2021 Mar 11. J Cancer Res Clin Oncol. 2021. PMID: 33693962 Free PMC article. - RNA binding protein-mediated competing endogenous RNA mechanism in cancer.
Zhu J, Mo YY, Peng WX. Zhu J, et al. Gene. 2025 Sep 5;963:149606. doi: 10.1016/j.gene.2025.149606. Epub 2025 Jun 4. Gene. 2025. PMID: 40480446 Review. - LncRNAOmics: A Comprehensive Review of Long Non-Coding RNAs in Plants.
Saha C, Saha S, Bhattacharyya NP. Saha C, et al. Genes (Basel). 2025 Jun 29;16(7):765. doi: 10.3390/genes16070765. Genes (Basel). 2025. PMID: 40725421 Free PMC article. Review.
Cited by
- Transcriptome Analysis of Subcutaneous Adipose Tissue from Severely Obese Patients Highlights Deregulation Profiles in Coding and Non-Coding Oncogenes.
Rey F, Messa L, Pandini C, Launi R, Barzaghini B, Micheletto G, Raimondi MT, Bertoli S, Cereda C, Zuccotti GV, Cancello R, Carelli S. Rey F, et al. Int J Mol Sci. 2021 Feb 17;22(4):1989. doi: 10.3390/ijms22041989. Int J Mol Sci. 2021. PMID: 33671464 Free PMC article. - Long Non-Coding RNAs in Liver Cancer and Nonalcoholic Steatohepatitis.
Uchida S, Kauppinen S. Uchida S, et al. Noncoding RNA. 2020 Aug 29;6(3):34. doi: 10.3390/ncrna6030034. Noncoding RNA. 2020. PMID: 32872482 Free PMC article. Review. - Characterisation and functional predictions of canine long non-coding RNAs.
Le Béguec C, Wucher V, Lagoutte L, Cadieu E, Botherel N, Hédan B, De Brito C, Guillory AS, André C, Derrien T, Hitte C. Le Béguec C, et al. Sci Rep. 2018 Sep 7;8(1):13444. doi: 10.1038/s41598-018-31770-2. Sci Rep. 2018. PMID: 30194329 Free PMC article. - LncRNA Tug1 relieves the steatosis of SelenoF-knockout hepatocytes via sponging miR-1934-3p.
Wang W, Miao Z, Qi X, Wang B, Liu Q, Shi X, Xu S. Wang W, et al. Cell Biol Toxicol. 2023 Dec;39(6):3175-3195. doi: 10.1007/s10565-023-09826-5. Epub 2023 Sep 18. Cell Biol Toxicol. 2023. PMID: 37721623 - Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre.
Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, Morse M, Engreitz J, Lander ES, Guttman M, Lodish HF, Flavell R, Raj A, Rinn JL. Hacisuleyman E, et al. Nat Struct Mol Biol. 2014 Feb;21(2):198-206. doi: 10.1038/nsmb.2764. Epub 2014 Jan 26. Nat Struct Mol Biol. 2014. PMID: 24463464 Free PMC article.
References
- Flegal KM, Carroll MD, Ogden CL, Curtin LR. Prevalence and trends in obesity among US adults, 1999-2008. JAMA. 2010;303(3):235–241. - PubMed
- Haslam DW, James WP. Obesity. Lancet. 2005;366(9492):1197–1209. - PubMed
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7(12):885–896. - PubMed
- Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14(11):1293–1307. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 GM099117/GM/NIGMS NIH HHS/United States
- DK068348/DK/NIDDK NIH HHS/United States
- R01 DK068348/DK/NIDDK NIH HHS/United States
- P50 HG006193/HG/NHGRI NIH HHS/United States
- P50HG006193-01/HG/NHGRI NIH HHS/United States
- DK047618/DK/NIDDK NIH HHS/United States
- P01 HL066105/HL/NHLBI NIH HHS/United States
- R37 DK047618/DK/NIDDK NIH HHS/United States
- 1DP2OD00667/OD/NIH HHS/United States
- P01GM099117/GM/NIGMS NIH HHS/United States
- R56 DK068348/DK/NIDDK NIH HHS/United States
- 5P01HL066105/HL/NHLBI NIH HHS/United States
- R01 DK047618/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases