Comparison of sequencing platforms for single nucleotide variant calls in a human sample - PubMed (original) (raw)
Comparison of sequencing platforms for single nucleotide variant calls in a human sample
Aakrosh Ratan et al. PLoS One. 2013.
Abstract
Next-generation sequencings platforms coupled with advanced bioinformatic tools enable re-sequencing of the human genome at high-speed and large cost savings. We compare sequencing platforms from Roche/454(GS FLX), Illumina/HiSeq (HiSeq 2000), and Life Technologies/SOLiD (SOLiD 3 ECC) for their ability to identify single nucleotide substitutions in whole genome sequences from the same human sample. We report on significant GC-related bias observed in the data sequenced on Illumina and SOLiD platforms. The differences in the variant calls were investigated with regards to coverage, and sequencing error. Some of the variants called by only one or two of the platforms were experimentally tested using mass spectrometry; a method that is independent of DNA sequencing. We establish several causes why variants remained unreported, specific to each platform. We report the indel called using the three sequencing technologies and from the obtained results we conclude that sequencing human genomes with more than a single platform and multiple libraries is beneficial when high level of accuracy is required.
Conflict of interest statement
Competing Interests: Some of the authors are employed by a commercial company, Genentech Inc. This does not alter the authors’ adherence to all the PLOS ONE policies on sharing data and materials.
Figures
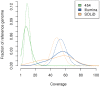
Figure 1. Depth of coverage distribution for the three platforms.
The y-axis indicates the fraction of the bases in the reference sequence that has a particular coverage. This does not include secondary alignments and potential PCR duplicates. The dashed lighter curves depict the coverage distribution as calculated using a Poisson model for each sequencing technology.
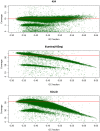
Figure 2. Variation of coverage with GC content in the three sequencing technologies.
The red line shows the mean coverage across the whole genome. Each point on the plot reflects the mean coverage and fraction of GC content in 50 kbp non-overlapping window. The y-axis shows the coverage whereas the x-axis shows the fraction of C, G nucleotides in the window. This does not include secondary alignments and potential PCR duplicates.
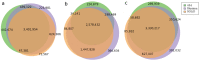
Figure 3. Venn diagram showing the overlap in the SNP calls made using data from the three sequencing technologies.
We display the sizes of each of the seven categories of overlaps among the variant calls in the three technologies. (a) depicts the overlaps when all substitution calls are used, (b) depicts the overlaps when all calls from Illumina and SOLiD are used but only the high-confidence subset of the 454 dataset is used, and (c) depicts the overlaps when only the variants in the uniquely alignable regions of the reference sequence are used.
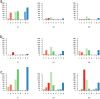
Figure 4. Discrepant SNP calls from each platform.
The categories on the x-axis are (1) no coverage at location (2) not enough coverage at location (3) more than expected coverage (4) alternate allele not seen (5) alternate allele seen just once (6) too many SNPs around location (7) close to a high-quality indel (8) low RMS mapping quality (9) low SNP quality. The y-axis depicts the number of locations (frequency) in each category. a) Comparison of SOLiD generated sequences with other sequences based on SNP calls and alignments. (i) SNPs called using 454 and Illumina sequences but not called using SOLiD reads. (ii) SNPs called only by SOLiD sequences. We investigate why they were not called using Illumina alignments. (iii) SNPs called only by SOLiD sequences. We investigate why they were not called using 454 alignments. b) Comparison of Illumina generated sequences with other sequences based on SNP calls and alignments. (i) SNPs called using 454 and SOLiD reads but not called using Illumina reads. (ii) SNPs called only by Illumina sequences. We investigate why they were not called using SOLiD alignments. (iii) SNPs called only by SOLiD sequences. We investigate why they were not called using 454 alignments. c) Comparison of 454 generated sequences with other sequences based on SNP calls and alignments. (i) SNPs called using SOLiD and Illumina reads but not called using 454 reads. (ii) SNPs called only by 454 sequences. We investigate why they were not called using SOLiD alignments. (iii) SNPs called only by 454 sequences. We investigate why they were not called using Illumina alignments.
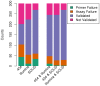
Figure 5. SNP Validation using Mass spectroscopy.
Validation of 300 putative SNP locations from each of the six sets of SNP calls in Figure 3a, where not all three technologies agree on the computed genotype. The categories on x-axis are “454” (SNPs called by 454 only), “Illumina” (SNPs called by Illumina only), “SOLiD” (SNPs called by SOLiD only), “454 & Illumina” (SNPs called by 454 and Illumina), “454 & SOLiD” (SNPs called by 454 and SOLiD), “Illumina & SOLiD” (SNPs called by Illumina and SOLiD). The color categories include “Primer Failure” (Primer extension failure), “Assay Failure” (Assay Failure), “Validated” and “Not Validated”.
Similar articles
- Comparison and evaluation of two exome capture kits and sequencing platforms for variant calling.
Zhang G, Wang J, Yang J, Li W, Deng Y, Li J, Huang J, Hu S, Zhang B. Zhang G, et al. BMC Genomics. 2015 Aug 5;16(1):581. doi: 10.1186/s12864-015-1796-6. BMC Genomics. 2015. PMID: 26242175 Free PMC article. - Germline and somatic variant identification using BGISEQ-500 and HiSeq X Ten whole genome sequencing.
Patch AM, Nones K, Kazakoff SH, Newell F, Wood S, Leonard C, Holmes O, Xu Q, Addala V, Creaney J, Robinson BW, Fu S, Geng C, Li T, Zhang W, Liang X, Rao J, Wang J, Tian M, Zhao Y, Teng F, Gou H, Yang B, Jiang H, Mu F, Pearson JV, Waddell N. Patch AM, et al. PLoS One. 2018 Jan 10;13(1):e0190264. doi: 10.1371/journal.pone.0190264. eCollection 2018. PLoS One. 2018. PMID: 29320538 Free PMC article. - Performance comparison of whole-genome sequencing platforms.
Lam HY, Clark MJ, Chen R, Chen R, Natsoulis G, O'Huallachain M, Dewey FE, Habegger L, Ashley EA, Gerstein MB, Butte AJ, Ji HP, Snyder M. Lam HY, et al. Nat Biotechnol. 2011 Dec 18;30(1):78-82. doi: 10.1038/nbt.2065. Nat Biotechnol. 2011. PMID: 22178993 Free PMC article. - Overview of Next-generation Sequencing Platforms Used in Published Draft Plant Genomes in Light of Genotypization of Immortelle Plant (Helichrysium Arenarium).
Hodzic J, Gurbeta L, Omanovic-Miklicanin E, Badnjevic A. Hodzic J, et al. Med Arch. 2017 Aug;71(4):288-292. doi: 10.5455/medarh.2017.71.288-292. Med Arch. 2017. PMID: 28974852 Free PMC article. Review. - Massively parallel sequencing approaches for characterization of structural variation.
Koboldt DC, Larson DE, Chen K, Ding L, Wilson RK. Koboldt DC, et al. Methods Mol Biol. 2012;838:369-84. doi: 10.1007/978-1-61779-507-7_18. Methods Mol Biol. 2012. PMID: 22228022 Free PMC article. Review.
Cited by
- Common copy number variation detection from multiple sequenced samples.
Duan J, Deng HW, Wang YP. Duan J, et al. IEEE Trans Biomed Eng. 2014 Mar;61(3):928-37. doi: 10.1109/TBME.2013.2292588. IEEE Trans Biomed Eng. 2014. PMID: 24557694 Free PMC article. - MSeq-CNV: accurate detection of Copy Number Variation from Sequencing of Multiple samples.
Malekpour SA, Pezeshk H, Sadeghi M. Malekpour SA, et al. Sci Rep. 2018 Mar 5;8(1):4009. doi: 10.1038/s41598-018-22323-8. Sci Rep. 2018. PMID: 29507384 Free PMC article. - The role of replicates for error mitigation in next-generation sequencing.
Robasky K, Lewis NE, Church GM. Robasky K, et al. Nat Rev Genet. 2014 Jan;15(1):56-62. doi: 10.1038/nrg3655. Epub 2013 Dec 10. Nat Rev Genet. 2014. PMID: 24322726 Free PMC article. Review. - Impact of next-generation sequencing error on analysis of barcoded plasmid libraries of known complexity and sequence.
Deakin CT, Deakin JJ, Ginn SL, Young P, Humphreys D, Suter CM, Alexander IE, Hallwirth CV. Deakin CT, et al. Nucleic Acids Res. 2014;42(16):e129. doi: 10.1093/nar/gku607. Epub 2014 Jul 10. Nucleic Acids Res. 2014. PMID: 25013183 Free PMC article. - Meta-analyses of studies of the human microbiota.
Lozupone CA, Stombaugh J, Gonzalez A, Ackermann G, Wendel D, Vázquez-Baeza Y, Jansson JK, Gordon JI, Knight R. Lozupone CA, et al. Genome Res. 2013 Oct;23(10):1704-14. doi: 10.1101/gr.151803.112. Epub 2013 Jul 16. Genome Res. 2013. PMID: 23861384 Free PMC article.
References
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, et al. (2001) The sequence of the human genome. Science (New York, NY) 291: 1304–1351 doi:10.1126/science.1058040. - DOI - PubMed
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 doi:10.1038/35057062. - DOI - PubMed
- Wheeler D a, Srinivasan M, Egholm M, Shen Y, Chen L, et al. (2008) The complete genome of an individual by massively parallel DNA sequencing. Nature 452: 872–876 doi:10.1038/nature06884. - DOI - PubMed
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, et al. (2007) The diploid genome sequence of an individual human. PLoS biology 5: e254 doi:10.1371/journal.pbio.0050254. - DOI - PMC - PubMed
- Schuster SC, Miller W, Ratan A, Tomsho LP, Giardine B, et al. (2010) Complete Khoisan and Bantu genomes from southern Africa. Nature 463: 943–947 doi:10.1038/nature08795. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous