DEAD-box helicases as integrators of RNA, nucleotide and protein binding - PubMed (original) (raw)
Review
DEAD-box helicases as integrators of RNA, nucleotide and protein binding
Andrea A Putnam et al. Biochim Biophys Acta. 2013 Aug.
Abstract
DEAD-box helicases perform diverse cellular functions in virtually all steps of RNA metabolism from Bacteria to Humans. Although DEAD-box helicases share a highly conserved core domain, the enzymes catalyze a wide range of biochemical reactions. In addition to the well established RNA unwinding and corresponding ATPase activities, DEAD-box helicases promote duplex formation and displace proteins from RNA. They can also function as assembly platforms for larger ribonucleoprotein complexes, and as metabolite sensors. This review aims to provide a perspective on the diverse biochemical features of DEAD-box helicases and connections to structural information. We discuss these data in the context of a model that views the enzymes as integrators of RNA, nucleotide, and protein binding. This article is part of a Special Issue entitled: The Biology of RNA helicases - Modulation for life.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures
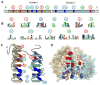
Figure 1. Architecture of the DEAD-box helicase core
(A) Schematic of the primary structure of the DEAD-box helicase core. Domain 1 (N-terminal, tan) and 2 (C-terminal, blue) designate the two RecA-like helicase domains. Circled numbers indicate the approximate location of the characteristic sequence motifs, colored according to their primary biochemical function (red, ATP binding and hydrolysis; blue, RNA binding, green, coordination between RNA and ATP binding). (B) Sequence conservation in the characteristic sequence motifs of DEAD-box helicases. Amino acid conservation is represented by the height of letter. Coloring marks the chemical properties of a given amino acid: green and purple — polar, blue — basic, red— acidic, and black— hydrophobic (adapted from ref. [1]). (C) Schematic representation the topology of the two RecA-like helicase core domains. β-strands are indicated by arrows, α-helices by cylinders. Characteristic sequence motifs are colored as in panel A (adapted from ref [1]). (D) Position of the characteristic sequence motifs in the three-dimensional structure of the DEAD-box helicase Vasa [19]. The bound ATP analog and Mg2+ ion are colored teal; the nucleic acid is colored orange. Sequence motifs are colored as in panels A to C.
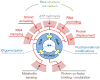
Figure 2. Biochemical features and activities of DEAD-box helicases
The inner circle marks the basic biochemical features, RNA, nucleotide (ATP) and protein binding. The yellow arrows mark the connection between the activities. The outer circle segments (light red) designate the activities that arise from either one or the coordination of two basic biochemical features. The red lines indicate which biochemical features contribute to a given activity. RNA structure conversion is based on the coordination between two activities, strand annealing and RNA unwinding, as indicated.
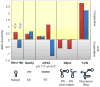
Figure 3. Cooperativity between RNA and ATP or ADP binding by DEAD-box RNA helicases
Cooperativity is expressed by changes in free binding energy [ΔΔG° (kcal/mol)] for ATP or ADP binding in the presence or absence of RNA. Values for ΔΔG° were calculated according to ΔΔG° = ΔG°(nt with RNA) − ΔG°(nt no RNA), with ΔG° = − RTlnKeq. Positive ΔΔG° values indicate cooperative binding of nucleotide and RNA, negative values indicate anti-cooperative binding. Values were calculated with published data for Mss116p, eIF4A, DbpA, YxiN [46,47,48,49,50], and for Ded1p with unpublished data (A.P. & E.J., unpublished observations). ΔΔG° = ΔG°(nt with RNA) − ΔG°(nt no RNA), with ΔG° = − RTlnKeq. RNAs used were as follows: Mss116p:28 nt RNA capable of forming a 12 bp hairpin with a 4 nt loop, Ded1p: 10 nt single stranded RNA, eIF4A: 21 nt single stranded RNA, DbpA: a 36 nt hairpin containing substrate with a single stranded region, or an additional 15 nt forming a second hairpin in place of the single stranded region, YxiN: a 154 nt structured substrate consisting of nucleotides 2481– 2634 of the B. subtilis 23S rRNA [46].
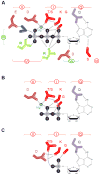
Figure 4. Schematic representation of nucleotide binding by DEAD-box proteins
Key residues in the helicase core domain that mediate ATP binding and hydrolysis, based on the structure of several DEAD-box proteins. Sequence motifs are colored and numbered as described in Fig. 1. (A) Binding of ATP with Mg2+. (B) Binding of ADP with Mg2+. (B) Binding of AMP.
Figure 5. Duplex unwinding by local strand separation
RNA strands are represented by black lines. Single-stranded region is shown in gray. The dotted line indicates requirement for proximity, but not physical connection to the duplex. The DEAD-box helicase is depicted by yellow and blue semi-circles representing the helicase N- and C-terminal domains respectively. More than 2 helicase protomers may participate in the unwinding for a subset of DEAD-box helicases, but as mentioned, DEAD-box helicases can also function as monomers. ATP is shown as red triangle. ATP hydrolysis and subsequent formation of ADP (purple triangle) and free phosphate is required for release of products and recycling of the enzyme. Dissociation of the helicase prior to complete strand separation results in an RNA species with a partially opened helix which will rapidly return to a duplex structure (adapted from refs. [7,91]).
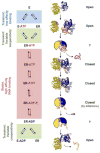
Figure 6. Structural basis for duplex unwinding by DEAD-box proteins
The left panel shows the scheme for the RNA-dependent ATP hydrolysis cycle. Relative RNA affinities for each stage of the cycle is indicated on the left of the scheme. The structures show each stage for which structural information is available, as discussed in the text. For stages without direct structural information, cartoons represent speculative structural arrangements. Structures depict in yellow the N-terminal domain and in blue the C-terminal domain. The following structures are shown: eIF4A without nucleotide or RNA (1FUU), N-terminal domain of DDX20 with ADPNP (3B7G), C-terminal domain of Mss116p with C-terminal extension (CTE, light blue) and duplex RNA (red and orange (4DB2), Mss116p with CTE (light blue), ADP-BeF3, Mg2+and single stranded RNA (orange) (3I61), Mss116p with CTE (light blue), ADP-AlF4, Mg2+, and single stranded RNA (3I62), and UAP56, ADP, and Mg2+ (1XTJ). On the right, the orientation of the helicase domains relative to each other is indicated for each stage. “?” indicates insufficient evidence for domain orientation.
Similar articles
- The DEAD box proteins DDX5 (p68) and DDX17 (p72): multi-tasking transcriptional regulators.
Fuller-Pace FV. Fuller-Pace FV. Biochim Biophys Acta. 2013 Aug;1829(8):756-63. doi: 10.1016/j.bbagrm.2013.03.004. Epub 2013 Mar 19. Biochim Biophys Acta. 2013. PMID: 23523990 Review. - Looking back on the birth of DEAD-box RNA helicases.
Linder P, Fuller-Pace FV. Linder P, et al. Biochim Biophys Acta. 2013 Aug;1829(8):750-5. doi: 10.1016/j.bbagrm.2013.03.007. Epub 2013 Mar 29. Biochim Biophys Acta. 2013. PMID: 23542735 Review. - Functions of DEAD-box proteins in bacteria: current knowledge and pending questions.
Iost I, Bizebard T, Dreyfus M. Iost I, et al. Biochim Biophys Acta. 2013 Aug;1829(8):866-77. doi: 10.1016/j.bbagrm.2013.01.012. Epub 2013 Feb 13. Biochim Biophys Acta. 2013. PMID: 23415794 Review. - Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p.
Mallam AL, Del Campo M, Gilman B, Sidote DJ, Lambowitz AM. Mallam AL, et al. Nature. 2012 Oct 4;490(7418):121-5. doi: 10.1038/nature11402. Epub 2012 Sep 2. Nature. 2012. PMID: 22940866 Free PMC article. - Duplex unwinding with DEAD-box proteins.
Jankowsky E, Putnam A. Jankowsky E, et al. Methods Mol Biol. 2010;587:245-64. doi: 10.1007/978-1-60327-355-8_18. Methods Mol Biol. 2010. PMID: 20225155
Cited by
- Modulation of RNA Condensation by the DEAD-Box Protein eIF4A.
Tauber D, Tauber G, Khong A, Van Treeck B, Pelletier J, Parker R. Tauber D, et al. Cell. 2020 Feb 6;180(3):411-426.e16. doi: 10.1016/j.cell.2019.12.031. Epub 2020 Jan 9. Cell. 2020. PMID: 31928844 Free PMC article. - The DEAD-box RNA helicase RhlE2 is a global regulator of Pseudomonas aeruginosa lifestyle and pathogenesis.
Hausmann S, Gonzalez D, Geiser J, Valentini M. Hausmann S, et al. Nucleic Acids Res. 2021 Jul 9;49(12):6925-6940. doi: 10.1093/nar/gkab503. Nucleic Acids Res. 2021. PMID: 34151378 Free PMC article. - Crystal structure, mutational analysis and RNA-dependent ATPase activity of the yeast DEAD-box pre-mRNA splicing factor Prp28.
Jacewicz A, Schwer B, Smith P, Shuman S. Jacewicz A, et al. Nucleic Acids Res. 2014 Nov 10;42(20):12885-98. doi: 10.1093/nar/gku930. Epub 2014 Oct 10. Nucleic Acids Res. 2014. PMID: 25303995 Free PMC article. - Cellular DEAD-box RNA helicase DDX6 modulates interaction of miR-122 with the 5' untranslated region of hepatitis C virus RNA.
Biegel JM, Henderson E, Cox EM, Bonenfant G, Netzband R, Kahn S, Eager R, Pager CT. Biegel JM, et al. Virology. 2017 Jul;507:231-241. doi: 10.1016/j.virol.2017.04.014. Epub 2017 Apr 26. Virology. 2017. PMID: 28456022 Free PMC article. - Biochemical Differences and Similarities between the DEAD-Box Helicase Orthologs DDX3X and Ded1p.
Sharma D, Putnam AA, Jankowsky E. Sharma D, et al. J Mol Biol. 2017 Nov 24;429(23):3730-3742. doi: 10.1016/j.jmb.2017.10.008. Epub 2017 Oct 13. J Mol Biol. 2017. PMID: 29037760 Free PMC article.
References
- Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. - PubMed
- Linder P. The Dynamic Life with DEAD-box RNA Helicases. In: Jankowsky E, editor. RNA Helicases: RSC Biomolecular Sciences. Vol. 19. 2010. pp. 32–60.
- Caruthers JM, McKay DB. Helicase structure and mechanism. Curr Opin Struct Biol. 2002;12:123–133. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases