Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential - PubMed (original) (raw)
. 2013 Feb 19;4(1):e00611-12.
doi: 10.1128/mBio.00611-12.
Hulda R Jónsdóttir, Doreen Muth, Ole J Hamming, Rune Hartmann, Regulo Rodriguez, Robert Geffers, Ron A M Fouchier, Christian Drosten, Marcel A Müller, Ronald Dijkman, Volker Thiel
Affiliations
- PMID: 23422412
- PMCID: PMC3573664
- DOI: 10.1128/mBio.00611-12
Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential
Eveline Kindler et al. mBio. 2013.
Abstract
The recent emergence of a novel human coronavirus (HCoV-EMC) in the Middle East raised considerable concerns, as it is associated with severe acute pneumonia, renal failure, and fatal outcome and thus resembles the clinical presentation of severe acute respiratory syndrome (SARS) observed in 2002 and 2003. Like SARS-CoV, HCoV-EMC is of zoonotic origin and closely related to bat coronaviruses. The human airway epithelium (HAE) represents the entry point and primary target tissue for respiratory viruses and is highly relevant for assessing the zoonotic potential of emerging respiratory viruses, such as HCoV-EMC. Here, we show that pseudostratified HAE cultures derived from different donors are highly permissive to HCoV-EMC infection, and by using reverse transcription (RT)-PCR and RNAseq data, we experimentally determined the identity of seven HCoV-EMC subgenomic mRNAs. Although the HAE cells were readily responsive to type I and type III interferon (IFN), we observed neither a pronounced inflammatory cytokine nor any detectable IFN responses following HCoV-EMC, SARS-CoV, or HCoV-229E infection, suggesting that innate immune evasion mechanisms and putative IFN antagonists of HCoV-EMC are operational in the new host. Importantly, however, we demonstrate that both type I and type III IFN can efficiently reduce HCoV-EMC replication in HAE cultures, providing a possible treatment option in cases of suspected HCoV-EMC infection. IMPORTANCE A novel human coronavirus, HCoV-EMC, has recently been described to be associated with severe respiratory tract infection and fatalities, similar to severe acute respiratory syndrome (SARS) observed during the 2002-2003 epidemic. Closely related coronaviruses replicate in bats, suggesting that, like SARS-CoV, HCoV-EMC is of zoonotic origin. Since the animal reservoir and circumstances of zoonotic transmission are yet elusive, it is critically important to assess potential species barriers of HCoV-EMC infection. An important first barrier against invading respiratory pathogens is the epithelium, representing the entry point and primary target tissue of respiratory viruses. We show that human bronchial epithelia are highly susceptible to HCoV-EMC infection. Furthermore, HCoV-EMC, like other coronaviruses, evades innate immune recognition, reflected by the lack of interferon and minimal inflammatory cytokine expression following infection. Importantly, type I and type III interferon treatment can efficiently reduce HCoV-EMC replication in the human airway epithelium, providing a possible avenue for treatment of emerging virus infections.
Figures
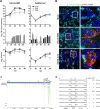
FIG 1
Replication of HCoV-EMC and SARS-CoV on HAE cultures. (A) HAE cultures from three donors (0712, black; 1001, light gray; 1505, dark gray) were prepared as described previously (7) and infected with HCoV-EMC or SARS-CoV (MOI = 0.1). Progeny virus release at the apical (top and bottom) and basolateral (middle) surfaces of HCoV-EMC- or SARS-CoV-infected HAE cultures was determined as genome equivalents (GE) or plaque-forming units (PFU) per ml at the indicated hpi by using quantitative real-time reverse transcription-PCR (qRT-PCR) specific for HCoV-EMC (16) and SARS-CoV (17) or titration of infectious particles on Vero cells. Experiments were performed in triplicate for each donor. Data are depicted as mean values ± standard deviations (SD); nd, not detected. (B) HCoV-EMC- and SARS-CoV-infected (MOI = 0.1) or mock-treated HAE cell cultures were fixed 48 hpi with 6% PFA and immunostained using the procedure as described (18). Rabbit polyclonal antiserum directed against SARS-CoV Nsp3 (green; anti-SARS-CoV antibody; Rockland) and mouse monoclonal antibody directed against dsRNA (red; J2; English & Scientific Consulting Bt.) were used as primary antibodies. Dylight 488-labeled anti-mouse IgG (H+L) and Dylight 647-labeled anti-rabbit IgG (H+L) (Jackson Immunoresearch) were applied as secondary antibodies, followed by two separate incubation steps with Cy3-conjugated mouse anti-β-tubulin antibody (light blue; Sigma) for staining of ciliated cells and DAPI (4',6-diamidino-2-phenylindole; Invitrogen) for staining nuclei (dark blue). Images were acquired using an EC, Plan-Neofluor 63×/1.40 oil differential inference contrast (DIC) M27 objective on a Zeiss LSM 710 confocal microscope. Image capture, analysis, and processing were performed using the ZEN 2010 (Zeiss) and Imaris (Bitplane Scientific Software) software packages. Representative images are shown from one (1505) of three donors. (C) Schematic representation of sequence reads of an RNAseq analysis of poly(A)-containing RNA derived from HCoV-EMC-infected HAE cultures (MOI = 1; 6 hpi). Single reads are depicted in green (sense) and red (antisense). The density of reads exceeding 34 for particular regions are shown condensed in gray. Blue arrows depict HCoV-EMC genes and open reading frames (ORFs). (D) Summary of detected HCoV-EMC mRNAs. Leader-body junctions of HCoV-EMC mRNAs are shown with 15 nucleotides upstream and downstream of the transcription regulatory sequence (TRS; bold). Numbers depict corresponding nucleotide positions in the HCoV-EMC genome. For all 8 viral mRNAs, the ORFs residing in the unique region and the method used for identification (RT-PCR or RNAseq) are indicated.
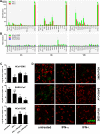
FIG 2
Human coronavirus-host interaction. (A) Gene expression analysis of IFN-treated HAE cultures. HAE cultures derived from three different donors were used untreated or were stimulated from the basolateral side with recombinant IFN-α (100 IU/ml; IFN-αA/D human; Sigma) or recombinant IFN-λ3 (10 ng/ml) (8) for 3, 6, and 12 h until total cellular RNA was extracted using RNeasy (Qiagen). Reverse transcription was performed with Moloney murine leukemia virus reverse transcriptase according to the manufacturer’s protocol (Invitrogen) using 1 µg of DNase-treated total RNA. Two microliters of diluted cDNA was amplified according to the manufacturer’s protocol, using primers targeting 15 different mRNA transcripts (see Table S1 in the supplemental material). Measurements and analysis were performed using a LightCycler 480 II instrument and software package (Roche). Cycle profile, 10 min at 95°C; 45 cycles of 10 s at 95°C, 20 s at 55°C, and 20 s at 72°C; followed by a melting curve step to confirm product specificity. Relative gene expression was calculated using the 2−ΔΔ_Ct_ method (19) and is shown as fold induction of IFN-treated samples compared to that of untreated controls. (B) Gene expression analysis of virus-infected HAE cultures. HAE cell cultures were infected with HCoV-EMC, SARS-CoV, or HCoV-229E (MOI = 1), and total cellular RNA was isolated at 3, 6, and 12 hpi. Relative gene expression analysis was performed as described above. (C) Analysis of virus replication following IFN pretreatment. HAE cell cultures were left untreated or were treated from the basolateral side for 16 h with recombinant IFN-α (100 IU/ml; Sigma) or recombinant IFN-λ3 (10 ng/ml or 100 ng/ml) (8). The basolateral medium was replaced prior to infection with HCoV-EMC, SARS-CoV, and HCoV-229E (MOI = 0.1). Apical progeny virus release was determined at 48 hpi by qRT-PCR and is given as GE per ml. Each bar represents the mean ± SD from independent experiments performed in duplicate using HAE cultures derived from three different donors. ns, not significant (P > 0.05); *, P < 0.05; **, P < 0.01 (paired t test). (D) Immunofluorescence analysis of IFN-treated and virus-infected HAE cultures. HAE cultures were fixed with 6% PFA and immunostained using the procedure as described (18). Mouse monoclonal antibody directed against dsRNA (J2; English & Scientific Consulting Bt.) was applied as primary antibody and Dylight 488-labeled anti-mouse IgG (H+L) as secondary antibody (green; Jackson ImmunoResearch), followed by staining of cilia with Cy3-conjugated mouse anti-β-tubulin antibody (red; Sigma). Images were acquired using an EC, Plan-Neofluor 63×/1.40 oil DIC M27 objective on a Zeiss 710 confocal laser scanning microscope. Image capture, analysis, and processing were performed using the ZEN 2010 (Zeiss) and Imaris (Bitplane Scientific Software) software packages. Representative images are shown from one (0401) of three donors.
Comment in
- The emergence of human coronavirus EMC: how scared should we be?
Chan RW, Poon LL. Chan RW, et al. mBio. 2013 Apr 9;4(2):e00191-13. doi: 10.1128/mBio.00191-13. mBio. 2013. PMID: 23572553 Free PMC article.
Similar articles
- Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines.
Müller MA, Raj VS, Muth D, Meyer B, Kallies S, Smits SL, Wollny R, Bestebroer TM, Specht S, Suliman T, Zimmermann K, Binger T, Eckerle I, Tschapka M, Zaki AM, Osterhaus AD, Fouchier RA, Haagmans BL, Drosten C. Müller MA, et al. mBio. 2012 Dec 11;3(6):e00515-12. doi: 10.1128/mBio.00515-12. mBio. 2012. PMID: 23232719 Free PMC article. - Tropism of and innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures.
Chan RW, Chan MC, Agnihothram S, Chan LL, Kuok DI, Fong JH, Guan Y, Poon LL, Baric RS, Nicholls JM, Peiris JS. Chan RW, et al. J Virol. 2013 Jun;87(12):6604-14. doi: 10.1128/JVI.00009-13. Epub 2013 Apr 3. J Virol. 2013. PMID: 23552422 Free PMC article. - Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans.
van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus AD, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RA. van Boheemen S, et al. mBio. 2012 Nov 20;3(6):e00473-12. doi: 10.1128/mBio.00473-12. mBio. 2012. PMID: 23170002 Free PMC article. - SARS-CoV replication and pathogenesis in an in vitro model of the human conducting airway epithelium.
Sims AC, Burkett SE, Yount B, Pickles RJ. Sims AC, et al. Virus Res. 2008 Apr;133(1):33-44. doi: 10.1016/j.virusres.2007.03.013. Epub 2007 Apr 23. Virus Res. 2008. PMID: 17451829 Free PMC article. Review. - Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS.
Drexler JF, Corman VM, Drosten C. Drexler JF, et al. Antiviral Res. 2014 Jan;101:45-56. doi: 10.1016/j.antiviral.2013.10.013. Epub 2013 Oct 31. Antiviral Res. 2014. PMID: 24184128 Free PMC article. Review.
Cited by
- MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment.
de Wilde AH, Raj VS, Oudshoorn D, Bestebroer TM, van Nieuwkoop S, Limpens RWAL, Posthuma CC, van der Meer Y, Bárcena M, Haagmans BL, Snijder EJ, van den Hoogen BG. de Wilde AH, et al. J Gen Virol. 2013 Aug;94(Pt 8):1749-1760. doi: 10.1099/vir.0.052910-0. Epub 2013 Apr 25. J Gen Virol. 2013. PMID: 23620378 Free PMC article. - Evaluation of alpaca tracheal explants as an ex vivo model for the study of Middle East respiratory syndrome coronavirus (MERS-CoV) infection.
Te N, Rodon J, Creve R, Pérez M, Segalés J, Vergara-Alert J, Bensaid A. Te N, et al. Vet Res. 2022 Sep 2;53(1):67. doi: 10.1186/s13567-022-01084-3. Vet Res. 2022. PMID: 36056449 Free PMC article. - Receptor for new coronavirus found.
Butler D. Butler D. Nature. 2013 Mar 14;495(7440):149-50. doi: 10.1038/495149a. Nature. 2013. PMID: 23486032 No abstract available. - Middle East Respiratory Syndrome Coronavirus ORF8b Accessory Protein Suppresses Type I IFN Expression by Impeding HSP70-Dependent Activation of IRF3 Kinase IKKε.
Wong LR, Ye ZW, Lui PY, Zheng X, Yuan S, Zhu L, Fung SY, Yuen KS, Siu KL, Yeung ML, Cai Z, Woo PC, Yuen KY, Chan CP, Jin DY. Wong LR, et al. J Immunol. 2020 Sep 15;205(6):1564-1579. doi: 10.4049/jimmunol.1901489. Epub 2020 Aug 3. J Immunol. 2020. PMID: 32747502 Free PMC article. - Human Organotypic Airway and Lung Organoid Cells of Bronchiolar and Alveolar Differentiation Are Permissive to Infection by Influenza and SARS-CoV-2 Respiratory Virus.
Ekanger CT, Zhou F, Bohan D, Lotsberg ML, Ramnefjell M, Hoareau L, Røsland GV, Lu N, Aanerud M, Gärtner F, Salminen PR, Bentsen M, Halvorsen T, Ræder H, Akslen LA, Langeland N, Cox R, Maury W, Stuhr LEB, Lorens JB, Engelsen AST. Ekanger CT, et al. Front Cell Infect Microbiol. 2022 Mar 14;12:841447. doi: 10.3389/fcimb.2022.841447. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35360113 Free PMC article.
References
- van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus AD, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RAM. 2012. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3(6):e00473-12 http://dx.doi.org/10.1128/mBio.00473-12 - PMC - PubMed
- Bermingham A, Chand MA, Brown CS, Aarons E, Tong C, Langrish C, Hoschler K, Brown K, Galiano M, Myers R, Pebody RG, Green HK, Boddington NL, Gopal R, Price N, Newsholme W, Drosten C, Fouchier RA, Zambon M. 2012. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 17:20290. - PubMed
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367:1814–1820 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous