Large chromosome deletions, duplications, and gene conversion events accumulate with age in normal human colon crypts - PubMed (original) (raw)
Large chromosome deletions, duplications, and gene conversion events accumulate with age in normal human colon crypts
John C F Hsieh et al. Aging Cell. 2013 Apr.
Abstract
Little is known about the types and numbers of mutations that may accumulate in normal human cells with age. Such information would require obtaining enough DNA from a single cell to accurately carry out reliable analysis despite extensive amplification; and complete genomic coverage under these circumstances is difficult. We have compared colon crypts, which are putatively clonal and contain ~2000 cells each, to determine how much somatic genetic variation occurs in vivo (without ex vivo cell culturing). Using high-density SNP microarrays, we find that chromosome deletions, duplications, and gene conversions were significantly more frequent in colons from the older individuals. These changes affected lengths ranging from 73 kb to 46 Mb. Although detection requires progeny of a single mutant stem cell to reach niche dominance over neighboring stem cells, none of the deletions appear likely to confer a selective advantage. Mutations can become fixed randomly during stem cell evolution through neutral drift in normal human crypts. The fact that chromosomal changes are detected in individual crypts with increasing age suggests that either such changes accumulate with age or single stem cell dominance increases with age, and the former is more likely. This progressive genome-wide divergence of human somatic cells with age has implications for aging and disease in multicellular organisms.
© 2013 Blackwell Publishing Ltd/Anatomical Society.
Figures
Figure 1. Colon Crypt Cell Dynamics and Detection of Chromosome Structural Alterations
Detection of chromosome changes in colon crypts depends on DSBs and repair in a stem cell, followed by niche succession such that mutant progeny are the majority of all crypt cells.
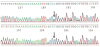
Figure 2. Locations of Chromosomal Structural Changes in Colons from Normal Individuals and Patients with Ulcerative Colitis
Black arrows indicate deletions, a black line indicates a duplication, and red lines indicate gene conversion by the breakage-induced replication mechanism.
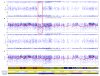
Figure 3. Deletion in One Colon Crypt That is Not Present in Other Crypts from the Same Individual
The SNP array results for individual 13 are shown for the 1.1 Mb deletion on chromosome 17. The top panel shows the allele frequency for heterozygous sites. Some SNP sites are BB (1.0 on this graph), some are AB (which are 0.5 on this graph), and some are AA (which is 0.0 on this graph). A deletion is detected as the red boxed region where there is a clear loss of the AB (heterozygous) status (loss of heterozygosity or LOH). Distinction between a deletion and a gene conversion event requires inspection of the Log R plot (second panel). The Log R plot is ideally 0.00 if both alleles are present, regardless of AA, AB or BB status. If only one allele is present (one missing), then the level falls to −1.0. If both alleles are missing, then the level falls to −2.0. A deletion manifests as a slight local decrease in the Log R plot at the same location as the loss of heterzygosity (LOH). (If there is no decrease in the Log R plot, then this region is a zone of gene conversion.) Comparison to three other colon crypts (only one is shown) from the same patient indicates that there is no LOH in the B allele frequency at this location (third panel from top) in the other crypts. The Log R plots of the three normal crypts also show no decrease at this location (fourth panel from top). The jagged horizontal red line in the Log R plots is a rolling average of the Log R ratio, and a persistent local dip indicates hemizygosity in the reigon of the LOH.
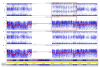
Figure 4. Long Gene Conversion Tract in One Colon Crypt That is Not Present in Other Crypts from the Same Individual
The SNP array results for individual 12, chromosome 17 show a 46 Mb LOH in one crypt. This is seen in the B allele frequency (top panel), which shows LOH from a point on the q arm of chromosome 17 to the telomere of that arm. This type of LOH is usually due to breakage-induced replication. The other crypts (only one shown) show no LOH in the B allele frequency.
Figure 5. Duplicated Region in One Colon Crypt That is Not Present in Other Crypts from the Same Individual
The results for sample e_9 (crypt 13a) from individual 13 showed not only the deletion on chromosome 5, but also a region of duplication on chromosome 9. The duplicated region is at 9q12–31.2 (bp position 65,629,772 to 109,557,941) is 44 Mb in length. The distinctive appearance of the B allele frequency plot identifies this region as having three copies: a double copy of one allele and a single copy of the other (otherwise, heterozygosity would be located at the 0.5 position, and homozygosity at the 1.0 or 0.0 position on the y-axis of the B allele frequency plot). The Log R plot is slightly elevated in the duplicated region, consistent with a duplication of one of the alleles.
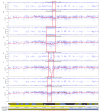
Figure 6. One Ulcerative Colitis Patient Has Four Out of Eight Crypts with Different Deletions in a Local Area of Chromosome 16p
The SNP array results show four different deletions, ranging from188 to 610 kb in size from individual #16. In one of these crypts (bottom two panels), both alleles are lost, as shown by the drop in the Log R plot focally to −2.0, and the B allele frequency in this region becomes unreliable because the Illumina software attempt to normalize the very small signals in this region. This accounts for the distribution of points from 0 to 1 in the B allele frequency of this crypt. The other three crypts simply show LOH (bottom six panels).
Figure 7. Double-Strand Breaks Can Explain Nearly All of the Large Chromosomal Changes Observed in Somatic Cells from Normal Individuals at Advanced Age
A. Breakage-Induced replication is thought to initiate from a DSB. One end of the DSB invades the homologous chromosome and copies to the end of that homologous chromosome. This mechanism accounts for loss of heterozygosity from a point to the end of the chromosome and where the entire region maintains the diploid copy number. B. Two DSBs can be rejoined in a manner that deletes the DNA between the two DSBs (portion in black). This mechanism accounts for loss of heterozygosity and a reduction in copy number from diploid to haploid. C. One DSB can invade a region of homology elsewhere on the same chromosome and delete the region of DNA between the two locations. This mechanism can also account for loss of heterozygosity with a reduction in copy number from diploid to haploid.
Figure 8. PCR-Based DNA Sequence Confirmation of Deletion in One Colon Crypt That is Not Present in Other Crypts from the Same Individual
The sequencing results of SNP rs4304440 on chromosome 9 for individual 14 are shown. The top panel displays the sequence from a normal crypt with heterozygosity at the given SNP position (upper black arrow). Note the double peak that is characteristic of the presence of both alleles. In contrast, the crypt with the LOH (bottom panel) displays homozygosity at the given SNP position (lower black arrow). This result confirms that the SNP is within the LOH region.
Similar articles
- Investigating stem cells in human colon by using methylation patterns.
Yatabe Y, Tavaré S, Shibata D. Yatabe Y, et al. Proc Natl Acad Sci U S A. 2001 Sep 11;98(19):10839-44. doi: 10.1073/pnas.191225998. Epub 2001 Aug 21. Proc Natl Acad Sci U S A. 2001. PMID: 11517339 Free PMC article. - High-resolution array copy number analyses for detection of deletion, gain, amplification and copy-neutral LOH in primary neuroblastoma tumors: four cases of homozygous deletions of the CDKN2A gene.
Carén H, Erichsen J, Olsson L, Enerbäck C, Sjöberg RM, Abrahamsson J, Kogner P, Martinsson T. Carén H, et al. BMC Genomics. 2008 Jul 29;9:353. doi: 10.1186/1471-2164-9-353. BMC Genomics. 2008. PMID: 18664255 Free PMC article. - 4p16.3 microdeletions and microduplications detected by chromosomal microarray analysis: New insights into mechanisms and critical regions.
Bi W, Cheung SW, Breman AM, Bacino CA. Bi W, et al. Am J Med Genet A. 2016 Oct;170(10):2540-50. doi: 10.1002/ajmg.a.37796. Epub 2016 Jun 10. Am J Med Genet A. 2016. PMID: 27287194 - Significance of genome-wide analysis of copy number alterations and UPD in myelodysplastic syndromes using combined CGH - SNP arrays.
Ahmad A, Iqbal MA. Ahmad A, et al. Curr Med Chem. 2012;19(22):3739-47. doi: 10.2174/092986712801661121. Curr Med Chem. 2012. PMID: 22680919 Review. - Methylation reveals a niche: stem cell succession in human colon crypts.
Kim KM, Shibata D. Kim KM, et al. Oncogene. 2002 Aug 12;21(35):5441-9. doi: 10.1038/sj.onc.1205604. Oncogene. 2002. PMID: 12154406 Review.
Cited by
- Aging and the rise of somatic cancer-associated mutations in normal tissues.
Risques RA, Kennedy SR. Risques RA, et al. PLoS Genet. 2018 Jan 4;14(1):e1007108. doi: 10.1371/journal.pgen.1007108. eCollection 2018 Jan. PLoS Genet. 2018. PMID: 29300727 Free PMC article. Review. - Cell-type-specific consequences of mosaic structural variants in hematopoietic stem and progenitor cells.
Grimes K, Jeong H, Amoah A, Xu N, Niemann J, Raeder B, Hasenfeld P, Stober C, Rausch T, Benito E, Jann JC, Nowak D, Emini R, Hoenicka M, Liebold A, Ho A, Shuai S, Geiger H, Sanders AD, Korbel JO. Grimes K, et al. Nat Genet. 2024 Jun;56(6):1134-1146. doi: 10.1038/s41588-024-01754-2. Epub 2024 May 28. Nat Genet. 2024. PMID: 38806714 Free PMC article. - Frequent Somatic Mutation in Adult Intestinal Stem Cells Drives Neoplasia and Genetic Mosaicism during Aging.
Siudeja K, Nassari S, Gervais L, Skorski P, Lameiras S, Stolfa D, Zande M, Bernard V, Frio TR, Bardin AJ. Siudeja K, et al. Cell Stem Cell. 2015 Dec 3;17(6):663-674. doi: 10.1016/j.stem.2015.09.016. Epub 2015 Oct 22. Cell Stem Cell. 2015. PMID: 26607382 Free PMC article. - The Initial Stage of Tumorigenesis in Drosophila Epithelial Tissues.
Tamori Y. Tamori Y. Adv Exp Med Biol. 2019;1167:87-103. doi: 10.1007/978-3-030-23629-8_5. Adv Exp Med Biol. 2019. PMID: 31520350 Review. - Clonal evolution of colorectal cancer in IBD.
Choi CR, Bakir IA, Hart AL, Graham TA. Choi CR, et al. Nat Rev Gastroenterol Hepatol. 2017 Apr;14(4):218-229. doi: 10.1038/nrgastro.2017.1. Epub 2017 Feb 8. Nat Rev Gastroenterol Hepatol. 2017. PMID: 28174420 Review.
References
- Bailey KJ, Maslov AY, Pruitt SC. Accumulation of mutations and somatic selection in aging neural stem/progenitor cells. Aging Cell. 2004;3:391–397. - PubMed
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. - PubMed
- Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C, Widaa S, Hinton J, Fahey C, Fu B, Swamy S, Dalgliesh GL, Teh BT, Deloukas P, Yang F, Campbell PJ, Futreal PA, Stratton MR. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. - PMC - PubMed
- Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous