Histone-methyltransferase MLL2 (KMT2B) is required for memory formation in mice - PubMed (original) (raw)
Histone-methyltransferase MLL2 (KMT2B) is required for memory formation in mice
Cemil Kerimoglu et al. J Neurosci. 2013.
Erratum in
- J Neurosci. 2013 Apr 17;33(16):7108
Abstract
The consolidation of long-term memories requires differential gene expression. Recent research has suggested that dynamic changes in chromatin structure play a role in regulating the gene expression program linked to memory formation. The contribution of histone methylation, an important regulatory mechanism of chromatin plasticity that is mediated by the counteracting activity of histone-methyltransferases and histone-demethylases, is, however, not well understood. Here we show that mice lacking the histone-methyltransferase myeloid/lymphoid or mixed-lineage leukemia 2 (mll2/kmt2b) gene in adult forebrain excitatory neurons display impaired hippocampus-dependent memory function. Consistent with the role of KMT2B in gene-activation DNA microarray analysis revealed that 152 genes were downregulated in the hippocampal dentate gyrus region of mice lacking kmt2b. Downregulated plasticity genes showed a specific deficit in histone 3 lysine 4 di- and trimethylation, while histone 3 lysine 4 monomethylation was not affected. Our data demonstrates that KMT2B mediates hippocampal histone 3 lysine 4 di- and trimethylation and is a critical player for memory formation.
Figures
Figure 1.
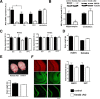
Mice lacking KMT2B from excitatory forebrain neurons show no overt phenotype. A, Loss of Kmt2b mRNA in the hippocampus and prefrontal cortex of Kmt2b cKO mice was confirmed via qPCR analysis. n = 9/group; **p < 0.01, ***p < 0.001, §p < 0.05. B, Immunoblot analysis shows that KMT2B protein levels are reduced in the hippocampus of kmt2b cKO mice (n = 4/group; *p < 0.05). C, qPCR analysis of kmt2a (mll1) and kmt2c (mll3) revealed no changes among kmt2b cKO and control mice (n = 5/group). D, Body weight of Kmt2b cKO and control mice was similar (n = 10/group). E, Brain weight of kmt2b cKO and control mice was similar (n = 8/group). F, Immunoreactivity for neuronal and synaptic marker proteins was similar in kmt2b cKO and control mice (n = 4/group). Left, Representative images. Scale bar, 300 μm. Right, Quantification (n = 4/group). DG, Dentate gyrus; CA, hippocampal CA region; PFC, prefrontal cortex; MAP2, microtubule-associated protein 2; SVP, synaptophysin; NeuN, Neuronal N. Error bars indicate SEM.
Figure 2.
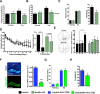
Mice lacking KMT2B shown impaired memory function. A, Mice were subjected to the novel object recognition paradigm. Wild-type mice (n = 12) and kmt2b cKO mice (n = 9) explored both objects similarly during the training session (left). When subjected to short-term memory test 5 min later, wild-type but not kmt2b cKO mice showed a significant preference for the novel object (*p < 0.05). B, When subjected to long-term memory version of the novel object recognition test both groups of mice showed a similar preference to the two identical objects during the training (left; control: n = 14, kmt2b cKO: n = 10). B, During the long-term memory test for object recognition control (n = 14) but not kmt2b cKO (n = 10) showed a preference for the novel object (right; ***p < 0.001). C, Left, Activity during the fear conditioning training and during the foot shock was similar among groups (Control: n = 20, kmt2b cKO: n = 15). Right, Freezing behavior was significantly impaired in kmt2b cKO mice when analyzed 24 h after the training (*p < 0.05). D, Escape latency during the water maze training was impaired in kmt2b cKO (n = 23) mice compared with the control group (n = 19) during the first and the last day of training (right). E, Left, Representative swim path during the probe test. Right, During the probe test only control (n = 19) but not kmt2b cKO (n = 23) mice showed a significant preference for the target quadrant (**p < 0.01). F, Four-month-old kmt2bfl/fl and wild-type mice were injected with AAV particles mediating CRE expression. Left, Representative images showing the expression of CRE-GFP in the dentate gyrus. The corresponding DAPI staining is shown for orientation. Scale bar, 500 μm. Right, Another group of mice was injected with the AAV-CRE virus (Wild-type: n = 6, Kmt2bfl/fl: n = 3) and dorsal dentate gyrus tissue was isolated 2 weeks later. qPCR analysis revealed the deletion of Kmt2b 2 weeks after injection in AAV-CRE-injected kmt2bfl/fl mice. G, Four-month-old kmt2bfl/fl and wild-type mice were injected with AAV-CRE and subjected to contextual fear conditioning 2 weeks after (n = 9/group). Deletion of kmt2b in the dentate gyrus of 4 month-old kmt2bfl/fl mice did not affect activity during the training and during the electric footshock (ES). H, Contextual freezing was impaired in kmt2bfl/fl mice compared with wild-type mice (*p < 0.05), indicative of disrupted memory formation. Error bars indicate SEM.
Figure 3.
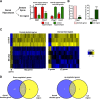
Loss of kmt2b leads to a deregulation of hippocampal transcriptome. A, RNA from the dorsal dentate gyrus and CA region was isolated. Specificity was confirmed by qPCR analysis for Wfs1 and Fmo1, two genes enriched in the dorsal hippocampus, and Plagl1 and Coch, two genes enriched in the ventral hippocampus (n = 4/group, **p < 0.01, ***p < 0.001). B, The dentate gyrus specificity was confirmed by qPCR analysis of the dentate gyrus-specific gene Tdo2 and the CA region-specific gene Lphn2 (n = 4/group; ***p < 0.001, **p < 001). C, Heat map showing differential gene expression among kmt2b cKO (n = 5) and control mice (n = 3) in the dorsal dentate gyrus and dorsal CA region. D, Venn diagram depicting downregulated and upregulated genes in the dorsal dentate gyrus and dorsal CA regions. Error bars indicate SEM.
Figure 4.
Downregulation of genes in kmt2b cKO mice is linked to altered H3K4 di- and trimethylation. A, Differential expression of selected genes identified in the gene array was confirmed via qPCR in an independent experiment (n = 4/group; ***p < 0.001, **p < 0.01, *p < 0.05 vs control). B–D, H3K4me3 (B), H3K4m2 (C), and H3K4me1 (D) levels for the selected genes was compared via ChIP analysis in kmt2b cKO and control mice (n = 4/group; ***p < 0.001, **p < 0.01, *p < 0.05 vs control). E, Three genes that were not altered in the gene array were used as control. Left, qPCR analysis confirmed similar expression of cFos, Egr-1, and Arc in Kmt2b cKO and control mice (n = 4/group). Right, ChIP analysis (n = 4/group) revealed that H3K4me3 at the TSS of c-Fos, Egr-1, and Arc genes was similar in kmt2b cKO and control mice. F, G, H3K9 acetylation (F) and H4K16 acetylation (G) levels for the selected genes were compared via ChIP analysis in Kmt2b cKO and control mice (n = 4/group; ***p < 0.001, **p < 0.01, *p < 0.001 vs control). Error bars indicate SEM.
Similar articles
- KMT2A and KMT2B Mediate Memory Function by Affecting Distinct Genomic Regions.
Kerimoglu C, Sakib MS, Jain G, Benito E, Burkhardt S, Capece V, Kaurani L, Halder R, Agís-Balboa RC, Stilling R, Urbanke H, Kranz A, Stewart AF, Fischer A. Kerimoglu C, et al. Cell Rep. 2017 Jul 18;20(3):538-548. doi: 10.1016/j.celrep.2017.06.072. Cell Rep. 2017. PMID: 28723559 - The histone methyltransferase KMT2B is required for RNA polymerase II association and protection from DNA methylation at the MagohB CpG island promoter.
Ladopoulos V, Hofemeister H, Hoogenkamp M, Riggs AD, Stewart AF, Bonifer C. Ladopoulos V, et al. Mol Cell Biol. 2013 Apr;33(7):1383-93. doi: 10.1128/MCB.01721-12. Epub 2013 Jan 28. Mol Cell Biol. 2013. PMID: 23358417 Free PMC article. - Neuronal Kmt2a/Mll1 histone methyltransferase is essential for prefrontal synaptic plasticity and working memory.
Jakovcevski M, Ruan H, Shen EY, Dincer A, Javidfar B, Ma Q, Peter CJ, Cheung I, Mitchell AC, Jiang Y, Lin CL, Pothula V, Stewart AF, Ernst P, Yao WD, Akbarian S. Jakovcevski M, et al. J Neurosci. 2015 Apr 1;35(13):5097-108. doi: 10.1523/JNEUROSCI.3004-14.2015. J Neurosci. 2015. PMID: 25834037 Free PMC article. - Structure, Activity and Function of the MLL2 (KMT2B) Protein Lysine Methyltransferase.
Klonou A, Chlamydas S, Piperi C. Klonou A, et al. Life (Basel). 2021 Aug 12;11(8):823. doi: 10.3390/life11080823. Life (Basel). 2021. PMID: 34440566 Free PMC article. Review. - COMPASS Ascending: Emerging clues regarding the roles of MLL3/KMT2C and MLL2/KMT2D proteins in cancer.
Fagan RJ, Dingwall AK. Fagan RJ, et al. Cancer Lett. 2019 Aug 28;458:56-65. doi: 10.1016/j.canlet.2019.05.024. Epub 2019 May 22. Cancer Lett. 2019. PMID: 31128216 Free PMC article. Review.
Cited by
- Generating new neurons to circumvent your fears: the role of IGF signaling.
Agis-Balboa RC, Fischer A. Agis-Balboa RC, et al. Cell Mol Life Sci. 2014 Jan;71(1):21-42. doi: 10.1007/s00018-013-1316-2. Epub 2013 Mar 30. Cell Mol Life Sci. 2014. PMID: 23543251 Free PMC article. Review. - KMT2B Is Selectively Required for Neuronal Transdifferentiation, and Its Loss Exposes Dystonia Candidate Genes.
Barbagiovanni G, Germain PL, Zech M, Atashpaz S, Lo Riso P, D'Antonio-Chronowska A, Tenderini E, Caiazzo M, Boesch S, Jech R, Haslinger B, Broccoli V, Stewart AF, Winkelmann J, Testa G. Barbagiovanni G, et al. Cell Rep. 2018 Oct 23;25(4):988-1001. doi: 10.1016/j.celrep.2018.09.067. Cell Rep. 2018. PMID: 30355503 Free PMC article. - Role of primary aging hallmarks in Alzheimer´s disease.
Zhao J, Huai J. Zhao J, et al. Theranostics. 2023 Jan 1;13(1):197-230. doi: 10.7150/thno.79535. eCollection 2023. Theranostics. 2023. PMID: 36593969 Free PMC article. Review. - Characterization of SETD1A haploinsufficiency in humans and Drosophila defines a novel neurodevelopmental syndrome.
Kummeling J, Stremmelaar DE, Raun N, Reijnders MRF, Willemsen MH, Ruiterkamp-Versteeg M, Schepens M, Man CCO, Gilissen C, Cho MT, McWalter K, Sinnema M, Wheless JW, Simon MEH, Genetti CA, Casey AM, Terhal PA, van der Smagt JJ, van Gassen KLI, Joset P, Bahr A, Steindl K, Rauch A, Keller E, Raas-Rothschild A, Koolen DA, Agrawal PB, Hoffman TL, Powell-Hamilton NN, Thiffault I, Engleman K, Zhou D, Bodamer O, Hoefele J, Riedhammer KM, Schwaibold EMC, Tasic V, Schubert D, Top D, Pfundt R, Higgs MR, Kramer JM, Kleefstra T. Kummeling J, et al. Mol Psychiatry. 2021 Jun;26(6):2013-2024. doi: 10.1038/s41380-020-0725-5. Epub 2020 Apr 28. Mol Psychiatry. 2021. PMID: 32346159 - Deciphering the molecular profile of plaques, memory decline and neuron loss in two mouse models for Alzheimer's disease by deep sequencing.
Bouter Y, Kacprowski T, Weissmann R, Dietrich K, Borgers H, Brauß A, Sperling C, Wirths O, Albrecht M, Jensen LR, Kuss AW, Bayer TA. Bouter Y, et al. Front Aging Neurosci. 2014 Apr 16;6:75. doi: 10.3389/fnagi.2014.00075. eCollection 2014. Front Aging Neurosci. 2014. PMID: 24795628 Free PMC article.
References
- Adegbola A, Gao H, Sommer S, Browning M. A novel mutation in JARID1C/SMCX in a patient with autism spectrum disorder (ASD) Am J Med Genet A. 2008;146A:505–511. - PubMed
- Agis-Balboa RC, Arcos-Diaz D, Wittnam J, Govindarajan N, Blom K, Burkhardt S, Haladyniak U, Agbemenyah HY, Zovoilis A, Salinas-Riester G, Opitz L, Sananbenesi F, Fischer A. A hippocampal insulin-growth factor 2 pathway regulates the extinction of fear memories. EMBO J. 2011;30:4071–4083. - PMC - PubMed
- Alarcón JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. - PubMed
- Albensi BC, Mattson MP. Evidence for the involvement of TNF and NF-kappaB in hippocampal synaptic plasticity. Synapse. 2000;35:151–159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous