In vivo analysis of human nucleoporin repeat domain interactions - PubMed (original) (raw)
In vivo analysis of human nucleoporin repeat domain interactions
Songli Xu et al. Mol Biol Cell. 2013 Apr.
Abstract
The nuclear pore complex (NPC), assembled from ∼30 proteins termed nucleoporins (Nups), mediates selective nucleocytoplasmic trafficking. A subset of nucleoporins bear a domain with multiple phenylalanine-glycine (FG) motifs. As binding sites for transport receptors, FG Nups are critical in translocation through the NPC. Certain FG Nups are believed to associate via low-affinity, cohesive interactions to form the permeability barrier of the pore, although the form and composition of this functional barrier are debated. We used green fluorescent protein-Nup98/HoxA9 constructs with various numbers of repeats and also substituted FG domains from other nucleoporins for the Nup98 domain to directly compare cohesive interactions in live cells by fluorescence recovery after photobleaching (FRAP). We find that cohesion is a function of both number and type of FG repeats. Glycine-leucine-FG (GLFG) repeat domains are the most cohesive. FG domains from several human nucleoporins showed no interactions in this assay; however, Nup214, with numerous VFG motifs, displayed measurable cohesion by FRAP. The cohesive nature of a human nucleoporin did not necessarily correlate with that of its yeast orthologue. The Nup98 GLFG domain also functions in pore targeting through binding to Nup93, positioning the GLFG domain in the center of the NPC and supporting a role for this nucleoporin in the permeability barrier.
Figures
FIGURE 1:
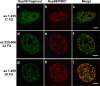
Association between Nup98 and leukemogenic Nup98 fusion proteins is a function of FG repeat number. GFP-Nup98 fragments (green) containing various numbers of nucleoporin FG repeat motifs (indicated as the total of all FG, FxFG, and GLFG motifs) were cotransfected with the CFP-tagged leukemogenic fusion protein Nup98/PMX1 (false colored in red) and visualized by confocal microscopy. The merged signal is shown at the right. Scale bars, 5 μm.
FIGURE 2:
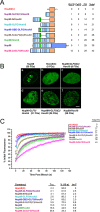
FG repeat number determines the affinity (cohesiveness?) of Nup98 fusion proteins. (A) Schematic of the different Nup98/HoxA9 constructs. FG repeats are indicated in light blue, GLFG repeats are in dark blue, and the single FXFG motif is in magenta. Repeats are positioned to scale, and the number of each type is indicated at right. (B) Localization of GFP-tagged exogenous Nup98, HoxA9 C-terminus, or Nup98/HoxA9 fusion proteins containing various numbers of repeats visualized by confocal microscopy. Number indicates total repeat motifs. Scale bar, 5 μm. (C) Dynamics of Nup98 fusion proteins with various numbers of FG motifs. GFP-tagged constructs were expressed in HeLa cells and photobleached, and fluorescence recovery was recorded over time. Curves are the average of at least six cells from two or more experiments. Error bars, SEM. Half-time of recovery, percent recovery at 10 s, and immobile fraction are indicated.
FIGURE 3:
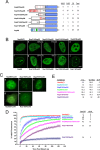
The contribution of the Nup98 repeat domain to NPC targeting is a function of the number of repeat motifs. (A) Schematic of the different Nup98 expression constructs. All constructs contain the full Nup98 C-terminal domain, which provides the major NPC-targeting motif. The number of each class of repeat motif is indicated at right. (B) Localization of GFP-tagged Nup98 constructs containing various numbers of repeats, as in A, visualized by confocal microscopy. Numbers indicate the total of all repeat motifs. Scale bar, 5 μm. (C) Quantification of NPC targeting. Low-expressing cells were chosen for analysis to minimize the formation of intranuclear bodies. Arrows indicate faint GLFG bodies seen at higher numbers of repeats. The relative nuclear rim intensity was scored as the ratio of nuclear rim intensity to intranuclear intensity in order to control for variation in cell-to-cell expression level. Between 9 and 35 individual cells were quantified for each construct as indicated. Statistically significant differences are indicated with p values. The relative nuclear rim intensity value for Nup98 with 39 repeats is somewhat inflated due to the strong propensity of nucleoplasmic protein to form GLFG bodies.
FIGURE 4:
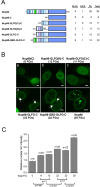
Repeat domains from non-GLFG nucleoporins are not functionally equivalent to Nup98 repeats. (A) Schematic of constructs containing FG repeat domains from different nucleoporins as indicated fused to the Nup98 C-terminus (dark blue). Charged amino acids within the repeat domains are indicated by red bars. The number of each class of nucleoporin repeat motif is indicated at the right. (B) Localization of GFP-tagged constructs from A by confocal microscopy. Nup214/Nup98 and Nup153/Nup98 each resulted in two classes of localization pattern (b and c, and g and h, respectively), and the percentage of cells exhibiting each pattern is given at the upper right. Scale bar, 5 μm. (C) Nucleoporin repeat domains as in A were fused to the HoxA9 C-terminus in place of Nup98. GFP-tagged constructs were localized by widefield microscopy. Scale bar, 5 μm. (D) FRAP recovery curves for repeat domains fused to the HoxA9 C-terminus. Nup214/HoxA9 and Nup153/HoxA9 curves represent FRAP of intranuclear and intranucleolar bodies, respectively. Curves are the average of 6 to 19 cells from two or more experiments. Data sets used for HoxA9(C), Nup98/HoxA9, and Nup98(2xGLFG)/HoxA9 are the same as those used in Figure 2. Error bars, SEM. (E) Quantitation of FRAP curves, as in Figure 2C.
FIGURE 5:
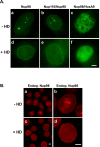
Interaction between GLFG repeats is sensitive to hexanediol. (A) Intranuclear GLFG bodies or Nup98/HoxA9 foci are dispersed by brief treatment of cells with HD. GFP-tagged proteins were transiently expressed and imaged by widefield microscopy before (a–c) or after (d–f) 30-s treatment with 5% HD. (B) Endogenous Nup98 GLFG bodies are dispersed by HD treatment. Nup98 in HeLa-C cells was detected by Nup98 antibody before (a, b) or after (c, d) HD treatment.
FIGURE 6:
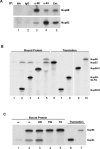
The Nup98 repeat domain interacts with Nup93 in a non–hexanediol-sensitive manner. (A) Nup98 and Nup93 can be coimmunoprecipitated. Nup98 or Nup93 was immunoprecipitated from Xenopus egg extract in the absence of detergent, and equal amounts of immunoprecipitate were analyzed by immunoblotting with antibodies to Nup98 or Nup93. (B) Nup93 binds to the repeat domains of Nup98. In vitro–translated Nup93 was incubated with unprogrammed translation reaction (lane 1), myc-tagged Nup98 fragments (Nup98-N, amino acids [aa] 1–225, lane 2; Nup98-GLFG, aa 220–506, lane 3; Nup98-C, aa 506–920, lane 4) or myc-tagged Nup98 (lane 5), and complexes were isolated on myc antibody beads and assessed by PAGE and autoradiography. Binding assays were carried out in the absence of detergent, leading to slight Nup93 background; however, Nup93 was significantly enriched on Nup98 or Nup98 fragments containing nucleoporin repeats (lanes 2, 3, and 5). (C) Binding of Nup93 to Nup98 is resistant to HD but sensitive to detergents. Binding assays were carried out as in B, using Nup93 alone (lane 1) or Nup93 together with myc-Nup98 (lanes 2–5). Bound proteins were isolated using myc antibody beads and washed well with buffer (lanes 1 and 2) or with buffer followed by one wash with 5% HD, 0.2% Tween-20 (TW, lane 4), or 0.2% TX-100 (TX, lane 5). Bound proteins were analyzed by PAGE and autoradiography.
Similar articles
- Intramolecular cohesion of coils mediated by phenylalanine--glycine motifs in the natively unfolded domain of a nucleoporin.
Krishnan VV, Lau EY, Yamada J, Denning DP, Patel SS, Colvin ME, Rexach MF. Krishnan VV, et al. PLoS Comput Biol. 2008 Aug 8;4(8):e1000145. doi: 10.1371/journal.pcbi.1000145. PLoS Comput Biol. 2008. PMID: 18688269 Free PMC article. - Entry into the nuclear pore complex is controlled by a cytoplasmic exclusion zone containing dynamic GLFG-repeat nucleoporin domains.
Fiserova J, Spink M, Richards SA, Saunter C, Goldberg MW. Fiserova J, et al. J Cell Sci. 2014 Jan 1;127(Pt 1):124-36. doi: 10.1242/jcs.133272. Epub 2013 Oct 21. J Cell Sci. 2014. PMID: 24144701 - Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex.
Patel SS, Belmont BJ, Sante JM, Rexach MF. Patel SS, et al. Cell. 2007 Apr 6;129(1):83-96. doi: 10.1016/j.cell.2007.01.044. Cell. 2007. PMID: 17418788 - Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport.
Terry LJ, Wente SR. Terry LJ, et al. Eukaryot Cell. 2009 Dec;8(12):1814-27. doi: 10.1128/EC.00225-09. Epub 2009 Oct 2. Eukaryot Cell. 2009. PMID: 19801417 Free PMC article. Review. - Interaction of nucleoporins with nuclear transport receptors: a structural perspective.
Kehlenbach RH, Neumann P, Ficner R, Dickmanns A. Kehlenbach RH, et al. Biol Chem. 2023 May 22;404(8-9):791-805. doi: 10.1515/hsz-2023-0155. Print 2023 Jul 26. Biol Chem. 2023. PMID: 37210735 Review.
Cited by
- Spelling out the roles of individual nucleoporins in nuclear export of mRNA.
Tingey M, Li Y, Yu W, Young A, Yang W. Tingey M, et al. Nucleus. 2022 Dec;13(1):170-193. doi: 10.1080/19491034.2022.2076965. Nucleus. 2022. PMID: 35593254 Free PMC article. Review. - Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors.
Andersen KR, Onischenko E, Tang JH, Kumar P, Chen JZ, Ulrich A, Liphardt JT, Weis K, Schwartz TU. Andersen KR, et al. Elife. 2013 Jun 11;2:e00745. doi: 10.7554/eLife.00745. Elife. 2013. PMID: 23795296 Free PMC article. - Biomolecular condensation of NUP98 fusion proteins drives leukemogenic gene expression.
Terlecki-Zaniewicz S, Humer T, Eder T, Schmoellerl J, Heyes E, Manhart G, Kuchynka N, Parapatics K, Liberante FG, Müller AC, Tomazou EM, Grebien F. Terlecki-Zaniewicz S, et al. Nat Struct Mol Biol. 2021 Feb;28(2):190-201. doi: 10.1038/s41594-020-00550-w. Epub 2021 Jan 21. Nat Struct Mol Biol. 2021. PMID: 33479542 Free PMC article. - Nuclear RNA-related processes modulate the assembly of cytoplasmic RNA granules.
Angel M, Fleshler E, Atrash MK, Kinor N, Benichou JIC, Shav-Tal Y. Angel M, et al. Nucleic Acids Res. 2024 May 22;52(9):5356-5375. doi: 10.1093/nar/gkae119. Nucleic Acids Res. 2024. PMID: 38366783 Free PMC article. - Disordered proteinaceous machines.
Fuxreiter M, Tóth-Petróczy Á, Kraut DA, Matouschek A, Lim RY, Xue B, Kurgan L, Uversky VN. Fuxreiter M, et al. Chem Rev. 2014 Jul 9;114(13):6806-43. doi: 10.1021/cr4007329. Epub 2014 Apr 4. Chem Rev. 2014. PMID: 24702702 Free PMC article. Review. No abstract available.
References
- Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. - PubMed
- Bayliss R, Littlewood T, Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell. 2000;102:99–108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous