Epigenomic plasticity enables human pancreatic α to β cell reprogramming - PubMed (original) (raw)
. 2013 Mar;123(3):1275-84.
doi: 10.1172/JCI66514. Epub 2013 Feb 22.
Affiliations
- PMID: 23434589
- PMCID: PMC3582140
- DOI: 10.1172/JCI66514
Epigenomic plasticity enables human pancreatic α to β cell reprogramming
Nuria C Bramswig et al. J Clin Invest. 2013 Mar.
Abstract
Insulin-secreting β cells and glucagon-secreting α cells maintain physiological blood glucose levels, and their malfunction drives diabetes development. Using ChIP sequencing and RNA sequencing analysis, we determined the epigenetic and transcriptional landscape of human pancreatic α, β, and exocrine cells. We found that, compared with exocrine and β cells, differentiated α cells exhibited many more genes bivalently marked by the activating H3K4me3 and repressing H3K27me3 histone modifications. This was particularly true for β cell signature genes involved in transcriptional regulation. Remarkably, thousands of these genes were in a monovalent state in β cells, carrying only the activating or repressing mark. Our epigenomic findings suggested that α to β cell reprogramming could be promoted by manipulating the histone methylation signature of human pancreatic islets. Indeed, we show that treatment of cultured pancreatic islets with a histone methyltransferase inhibitor leads to colocalization of both glucagon and insulin and glucagon and insulin promoter factor 1 (PDX1) in human islets and colocalization of both glucagon and insulin in mouse islets. Thus, mammalian pancreatic islet cells display cell-type-specific epigenomic plasticity, suggesting that epigenomic manipulation could provide a path to cell reprogramming and novel cell replacement-based therapies for diabetes.
Figures
Figure 1. Study design for determination of the transcriptome and differential histone marks in sorted human islet cells.
(A) Human islets were dispersed and subjected to FACS to obtain cell populations highly enriched for α, β, and exocrine (duct and acinar) cells. Chromatin was prepared and precipitated with antibodies for H3K4me3 and H3K27me3 followed by high-throughput sequencing (ChIP-Seq) (H3K4me3: n = 4 α, n = 4 β, n = 2 exocrine, H3K27me3: n = 3 α, n = 3 β, n = 2 exocrine). RNA-Seq analysis was performed to determine mRNA and lncRNA levels (n = 3 α, n = 3 β, n = 2 exocrine). (B) Sample purity assessment. Normalized insulin and glucagon expression levels of the individual α and β cell populations were obtained by qRT-PCR to calculate the contamination by the opposite cell population, revealing high sample purity (2.5%–10.3% contamination in the α and 2-13.1% contamination in the β cell populations; details in Supplemental Methods). (C) Analysis pipeline for H3K4me3 and H3K27me3 ChIP-Seq data. Peak calling (H3K4me3: GLITR; H3K27me3: STAR) on individual replicates, followed by signal pooling, was employed to assess histone modification profiles of α, β, and exocrine cells. Heat map analysis confirmed reproducibility of replicates. (D) Genome browser image of the PDX1 locus showing H3K4me3 enrichment in α, β, and exocrine cells and H3K27me3 enrichment only in α cells (defined as monovalent H3K4me3 enrichment in β and exocrine cells, bivalent mark in α cells; CpG islands: red bars).
Figure 2. Genome-wide transcriptome analysis using RNA-Seq confirms high purity of sorted cell populations and reveals cell-type–specific gene expression.
(A) Principal component analysis displays distinct cell populations and clustering of replicates (n = 3 α, n = 3 β, n = 2 exocrine), which confirms the high purity of our sorted cell populations (dots, replicates; crosses, averages). (B) Heat map analysis shows groups of genes with distinct expression patterns across cell types (columns: cell types, rows: genes). The orange, blue, and yellow bars on the left side of the heat map indicate α, β, and exocrine cell–specific gene clusters, respectively. The darker portion of these bars indicates stronger cell-type specificity of the gene cluster. We highlight important genes, including genes found to be associated with diabetes in genome-wide association studies (marked with asterisks). The complete gene lists of α, β, and exocrine cell–specific genes are provided in Supplemental Table 2.
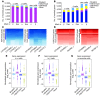
Figure 3. Human α, β, and exocrine cells exhibit convergent monovalent H3K4me3 and H3K27me3 profiles, which correlate highly with genome-wide expression data.
(A) The majority of H3K4me3-marked genes are shared between α, β, and exocrine cells (their overlap is indicated in the purple portion of the bars, 83%–95%). (B) H3K27me3 modification patterns are similar among pancreatic cell types (73%–83%, dark blue portion of the bars). (C and D) Heat map analysis (columns, individual samples; rows, genes) confirms low interindividual variability for all H3K4me3 (C) and H3K27me3 (D) peaks identified from the pooled data (peaks called by algorithms are indicated by the solid bars on the left of the heat maps). All pairs of columns in every heat map are significantly correlated based on correlation t test assessed by R statistics software (P < 2.2 × 10–16). (E–G) Normalized expression values obtained by RNA-Seq for genes grouped by their histone modification status in each cell type are shifted significantly above or below baseline expression (Wilcoxon signed rank test, P < 2.2 × 10–16). A shift above 0 on this scale indicates highly expressed genes and was observed for gene groups marked solely by H3K4me3 in all cell types (pink boxes in E–G). A shift below 0 on this scale indicates low or nonexpressed genes and was observed in all bivalently marked gene groups (light blue boxes) and monovalently H3K27me3-marked genes (dark blue boxes) in all cell types. Therefore, the histone modification states are significantly correlated with gene expression levels.
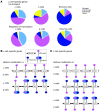
Figure 4. Human α cells demonstrate a higher number of bivalently marked genes than β and exocrine cells.
(A) Here, α cells display more bivalently marked loci than β and exocrine cells. Nearly half of the genes bivalently marked in α cells carry a monovalent mark in β cells (purple and dark blue portion of the far left bar corresponding to H3K4me3 and H3K27me3 marks in β cells, respectively). (B) 406 genes are marked bivalently in β cells, but monovalently by H3K4me3 in α cells, and gene ontology analysis for these genes shows 3 modestly enriched categories: regulation (reg.) of RNA metabolic process, regulation of transcription, and transcription. (C) Genes marked bivalently in α cells, but monovalently by H3K27me3 in β cells, are significantly enriched for developmental processes. For detailed GO analysis see Supplemental Table 6. (D) Comparison of transcriptional regulators marked bivalently in hESC (22) to the histone modification signatures of human α and β cells reveals a higher overlap between α cells and hESCs (44%, right pie chart) than between β cells and hESC (26%, left pie chart). Many of the genes marked bivalently both in α cells and hESCs carry the repressive mark in β cells (43%, dark blue portion of inset).
Figure 5. Human α cells display higher bivalency in genes encoding β cell transcriptional regulatory proteins.
(A) The epigenetic status of β cell signature genes (Figure 2B) functioning in ion transport or regulation of transcription was analyzed separately for α, β, and exocrine cells. Of the β cell–enriched ion transport genes, only 6% and 15% were marked bivalently in exocrine and β cells, respectively, while 29% carried this mark in α cells. For β cell signature genes involved in transcriptional regulation, 42% were marked as bivalent in α cells, but only 16% and 13% in β cells and exocrine cells, respectively. Thus α cells display a higher degree of bivalency for genes important in transcriptional regulation than for genes implicated in ion transport. (B and C) Schematic representation of the histone modification status of a relevant subset of human α and β cell signature genes (Figure 2B). The histone modification status of β cells is shown below each gene of interest. (B) As expected, most α cell signature genes are marked monovalently by H3K4me3 in α cells, and many of them carry a monovalent H3K27me3 mark in β cells. Interestingly, IRX1 and ARX are marked bivalently in α cells. (C) Within this subgroup of genes, β cell–expressed genes are marked monovalently by H3K4me3 in β cells, with the exception of HDAC9, which is marked bivalently. Remarkably, many β cell–expressed genes are marked bivalently in α cells, including the crucial insulin-synthesis enzyme PCSK1, the GLP1-receptor (GLP1R), and 2 essential β cell–specific transcription factors, MAFA and PDX1.
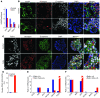
Figure 6. Inhibition of histone methyltransferases leads to partial endocrine cell-fate conversion.
(A) H3K27me3 ChIP-Seq analysis of human islets shows decreased H3K27me3 levels at the ARX, MAFA, and PDX1 loci following treatment of human islets with the histone methyltransferase inhibitor Adox. (B) Adox-treatment of human islets results in colocalization of glucagon (red) and insulin (green) granules within the same cell (yellow arrows), suggesting partial endocrine cell fate conversion, which was not seen in vehicle-treated islets (control). Original magnification, ×63. For Z-stack confocal images see Supplemental Videos 1 and 2. (C) Treatment of human islets with Adox results in colocalization of the β cell–specific transcription factor Pdx1 (white) and glucagon (red), further indicating endocrine reprogramming (white arrows: glucagon-positive, Pdx1-negative cells; yellow arrows: glucagon-positive, Pdx1-positive cells). The images on the right correspond to the area within yellow box. Original magnification, ×63. (D) Quantification of glucagon-positive, Pdx1-positive cells in untreated and Adox-treated human islets reveals many double-positive cells after Adox treatment, indicating initiation of reprogramming events in α cells. (E) Adox treatment of human islets leads to a decrease in NKX6-1 and MAFA levels in β cells (n = 3 α, n = 3 β, n = 2 treated α, n = 2 treated β), an increase in PDX1-levels, and no change in INS and GCG levels. (F) In Adox-treated α cells, we observe no change in INS and GCG expression, a slight decrease in NKX6-1 and MAFA levels, and an increase of ARX and PDX1 expression.
Comment in
- Creating new β cells: cellular transmutation by genomic alchemy.
Moss LG. Moss LG. J Clin Invest. 2013 Mar;123(3):1007-10. doi: 10.1172/JCI68348. Epub 2013 Feb 22. J Clin Invest. 2013. PMID: 23434598 Free PMC article.
Similar articles
- S6K1 controls epigenetic plasticity for the expression of pancreatic α/β cell marker genes.
Yi SA, Lee J, Park JW, Han J, Lee MG, Nam KH, Park JH, Oh H, Ahn SJ, Kim S, Kwon SH, Jo DG, Han JW. Yi SA, et al. J Cell Biochem. 2018 Aug;119(8):6674-6683. doi: 10.1002/jcb.26853. Epub 2018 Apr 17. J Cell Biochem. 2018. PMID: 29665055 - Genome-wide analysis of histone modifications in human pancreatic islets.
Bhandare R, Schug J, Le Lay J, Fox A, Smirnova O, Liu C, Naji A, Kaestner KH. Bhandare R, et al. Genome Res. 2010 Apr;20(4):428-33. doi: 10.1101/gr.102038.109. Epub 2010 Feb 24. Genome Res. 2010. PMID: 20181961 Free PMC article. - Creating new β cells: cellular transmutation by genomic alchemy.
Moss LG. Moss LG. J Clin Invest. 2013 Mar;123(3):1007-10. doi: 10.1172/JCI68348. Epub 2013 Feb 22. J Clin Invest. 2013. PMID: 23434598 Free PMC article. - Glucagon-producing α-cell transcriptional identity and reprogramming towards insulin production.
Oropeza D, Herrera PL. Oropeza D, et al. Trends Cell Biol. 2024 Mar;34(3):180-197. doi: 10.1016/j.tcb.2023.07.004. Epub 2023 Aug 23. Trends Cell Biol. 2024. PMID: 37626005 Review. - α-cell role in β-cell generation and regeneration.
Habener JF, Stanojevic V. Habener JF, et al. Islets. 2012 May-Jun;4(3):188-98. doi: 10.4161/isl.20500. Islets. 2012. PMID: 22847495 Free PMC article. Review.
Cited by
- Pancreatic Alpha-Cells Contribute Together With Beta-Cells to CXCL10 Expression in Type 1 Diabetes.
Nigi L, Brusco N, Grieco GE, Licata G, Krogvold L, Marselli L, Gysemans C, Overbergh L, Marchetti P, Mathieu C, Dahl Jørgensen K, Sebastiani G, Dotta F. Nigi L, et al. Front Endocrinol (Lausanne). 2020 Sep 15;11:630. doi: 10.3389/fendo.2020.00630. eCollection 2020. Front Endocrinol (Lausanne). 2020. PMID: 33042009 Free PMC article. - Advances in β cell replacement and regeneration strategies for treating diabetes.
Benthuysen JR, Carrano AC, Sander M. Benthuysen JR, et al. J Clin Invest. 2016 Oct 3;126(10):3651-3660. doi: 10.1172/JCI87439. Epub 2016 Oct 3. J Clin Invest. 2016. PMID: 27694741 Free PMC article. Review. - Ectonucleoside Triphosphate Diphosphohydrolase-3 Antibody Targets Adult Human Pancreatic β Cells for In Vitro and In Vivo Analysis.
Saunders DC, Brissova M, Phillips N, Shrestha S, Walker JT, Aramandla R, Poffenberger G, Flaherty DK, Weller KP, Pelletier J, Cooper T, Goff MT, Virostko J, Shostak A, Dean ED, Greiner DL, Shultz LD, Prasad N, Levy SE, Carnahan RH, Dai C, Sévigny J, Powers AC. Saunders DC, et al. Cell Metab. 2019 Mar 5;29(3):745-754.e4. doi: 10.1016/j.cmet.2018.10.007. Epub 2018 Nov 15. Cell Metab. 2019. PMID: 30449685 Free PMC article. - Overcoming the Limitations of Stem Cell-Derived Beta Cells.
Karimova MV, Gvazava IG, Vorotelyak EA. Karimova MV, et al. Biomolecules. 2022 Jun 9;12(6):810. doi: 10.3390/biom12060810. Biomolecules. 2022. PMID: 35740935 Free PMC article. Review. - Polycomb Repressive Complexes: Shaping Pancreatic Beta-Cell Destiny in Development and Metabolic Disease.
Varghese SS, Dhawan S. Varghese SS, et al. Front Cell Dev Biol. 2022 May 4;10:868592. doi: 10.3389/fcell.2022.868592. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35602600 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01DK088383/DK/NIDDK NIH HHS/United States
- U01 DK089529/DK/NIDDK NIH HHS/United States
- U01 DK089569/DK/NIDDK NIH HHS/United States
- U01DK089569/DK/NIDDK NIH HHS/United States
- R01 DK088383/DK/NIDDK NIH HHS/United States
- U42 RR006042/RR/NCRR NIH HHS/United States
- U01DK089529/DK/NIDDK NIH HHS/United States
- U01 DK070430/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases