Annotation and structural elucidation of bovine milk oligosaccharides and determination of novel fucosylated structures - PubMed (original) (raw)
Comparative Study
Annotation and structural elucidation of bovine milk oligosaccharides and determination of novel fucosylated structures
Danielle L Aldredge et al. Glycobiology. 2013 Jun.
Abstract
Bovine milk oligosaccharides (BMOs) are recognized by the dairy and food industries, as well as by infant formula manufacturers, as novel, high-potential bioactive food ingredients. Recent studies revealed that bovine milk contains complex oligosaccharides structurally related to those previously thought to be present in only human milk. These BMOs are microbiotic modulators involved in important biological activities, including preventing pathogen binding to the intestinal epithelium and serving as nutrients for a selected class of beneficial bacteria. Only a small number of BMO structures are fully elucidated. To better understand the potential of BMOs as a class of biotherapeutics, their detailed structure analysis is needed. This study initiated the development of a structure library of BMOs and a comprehensive evaluation of structure-related specificity. The bovine milk glycome was profiled by high-performance mass spectrometry and advanced separation techniques to obtain a comprehensive catalog of BMOs, including several novel, lower abundant neutral and fucosylated oligosaccharides that are often overlooked during analysis. Structures were identified using isomer-specific tandem mass spectroscopy and targeted exoglycosidase digestions to produce a BMO library detailing retention time, accurate mass and structure to allow their rapid identification in future studies.
Keywords: bovine colostrum; high-performance liquid chromatography; oligosaccharides; tandem mass spectrometry.
Figures
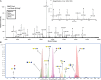
Fig. 1.
(A) The MALDI FT-ICR MS profile of a reduced BMO pool in the positive-ion mode. The numbers shown above the peaks represent, from bottom to top, the number of Hex, HexNAc, Fuc and NeuAc residues. Most ions are [M+Na]+ and the [M-H+2Na]+ ions are denoted with a star. (B) The -HPLC-Chip/TOF MS profile of the reduced BMO pool. Major peaks are labeled with putative structures, where blue circles denote Glc, blue squares denote GlcNAc, yellow circles denote Gal, purple diamonds denote NeuAc and red triangles denote Fuc. (These symbols are used throughout all figures.)
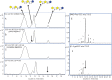
Fig. 2.
(A) MALDI FT-ICR MS profile of fraction 28 (from the off-line BMO pool separation) showing the isolation of m/z 1097.4 with EIC inset. (B) MS profile of fraction 28 after digestion with β1-4 galactosidase showing loss of two hexose mass shift with the EIC inset.
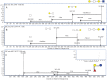
Fig. 3.
(A) EIC of m/z 507.2 from the BMO pool showing four isomers with their respective structures labeled. (B) EIC of m/z 507.2 at the half-way point of digestion with β1-3,6 galactosidase. (C) EIC of m/z 507.2 after full-time digestion with β1-3,6 galactosidase. (D) EIC of m/z 507.2 after digestion with β1-4 galactosidase. (E) EIC of m/z 710.2 from the BMO pool showing seven isomers. (F) EIC of m/z 710.2 after digestion with β1-4 galactosidase.
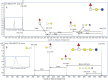
Fig. 4.
(A) EIC of m/z 548.2 from the BMO pool showing two main isomers. (B) EIC of m/z 548.2 after digestion with α _N_-acetylgalactosaminidase. (C) EIC of m/z 548.2 after digestion with β _N_-acetylhexosaminidase. (D) Magnification on base line of m/z 548.2 from the BMO pool showing additional isomers. (E) Isomer-specific fragmentation of EIC m/z 548.2 at 12.5 min, confirming reducing end HexNAc.
Fig. 5.
(A) Isomer-specific fragmentation profile of EIC m/z 751.3 from the BMO pool at 14.8 min, with the putative structure inset. (B) Isomer-specific fragmentation profile of EIC m/z 751.3 from the BMO pool at 14.1 min, with the putative structure inset. (C) Isomer-specific fragmentation profile of EIC m/z 491.19 from the BMO pool at 12.6 min, with the structure and EIC inset.
Fig. 6.
(A) Isomer-specific fragmentation of EIC m/z 694.3 at 10.4 min, with the putative structure inset. (B) Isomer-specific fragmentation of EIC m/z 1018.4 at 14.3 min, with the putative structure inset.
Fig. 7.
(A) Isomer-specific fragmentation of EIC m/z 1059.4 at 13.8 min, with the putative structure inset. (B) BPC of the BMO pool. (C) BPC of the HMO pool. (D) EIC of m/z 507.2 from the BMO pool showing four isomers. (E) EIC of m/z 507.2 from the HMO pool showing two isomers.
Similar articles
- Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.
Ryan R, Hill S. Ryan R, et al. Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article. - Genedrive kit for detecting single nucleotide polymorphism m.1555A>G in neonates and their mothers: a systematic review and cost-effectiveness analysis.
Shabaninejad H, Kenny RP, Robinson T, Stoniute A, O'Keefe H, Still M, Thornton C, Pearson F, Beyer F, Meader N. Shabaninejad H, et al. Health Technol Assess. 2024 Oct;28(75):1-75. doi: 10.3310/TGAC4201. Health Technol Assess. 2024. PMID: 39487741 Free PMC article. - Enabling Systemic Identification and Functionality Profiling for Cdc42 Homeostatic Modulators.
Malasala S, Azimian F, Chen YH, Twiss JL, Boykin C, Akhtar SN, Lu Q. Malasala S, et al. bioRxiv [Preprint]. 2024 Jan 8:2024.01.05.574351. doi: 10.1101/2024.01.05.574351. bioRxiv. 2024. PMID: 38260445 Free PMC article. Updated. Preprint. - Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.
Triana L, Palacios Huatuco RM, Campilgio G, Liscano E. Triana L, et al. Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
- Safety of 2'-fucosyllactose (2'-FL) produced by a derivative strain (Escherichia coli SGR5) of E. coli W (ATCC 9637) as a Novel Food pursuant to Regulation (EU) 2015/2283.
EFSA Panel on Nutrition, Novel Foods and Food Allergens (NDA); Turck D, Bohn T, Castenmiller J, De Henauw S, Hirsch-Ernst KI, Maciuk A, Mangelsdorf I, McArdle HJ, Naska A, Pentieva K, Siani A, Thies F, Tsabouri S, Vinceti M, Aguilera-Gómez M, Cubadda F, Frenzel T, Heinonen M, Prieto Maradona M, Marchelli R, Neuhäuser-Berthold M, Peláez C, Poulsen M, Schlatter JR, Siskos A, van Loveren H, Colombo P, Noriega Fernández E, Knutsen HK. EFSA Panel on Nutrition, Novel Foods and Food Allergens (NDA), et al. EFSA J. 2023 Nov 14;21(11):e08333. doi: 10.2903/j.efsa.2023.8333. eCollection 2023 Nov. EFSA J. 2023. PMID: 38027450 Free PMC article. - An improved method for the purification of milk oligosaccharides by graphitised carbon-solid phase extraction.
Robinson RC, Colet E, Tian T, Poulsen NA, Barile D. Robinson RC, et al. Int Dairy J. 2018 May;80:62-68. doi: 10.1016/j.idairyj.2017.12.009. Epub 2018 Jan 9. Int Dairy J. 2018. PMID: 30057440 Free PMC article. - New strategies for profiling and characterization of human milk oligosaccharides.
Porfirio S, Archer-Hartmann S, Moreau GB, Ramakrishnan G, Haque R, Kirkpatrick BD, Petri WA, Azadi P. Porfirio S, et al. Glycobiology. 2020 Sep 28;30(10):774-786. doi: 10.1093/glycob/cwaa028. Glycobiology. 2020. PMID: 32248230 Free PMC article. - Characterization of goat milk lactoferrin N-glycans and comparison with the N-glycomes of human and bovine milk.
Le Parc A, Dallas DC, Duaut S, Leonil J, Martin P, Barile D. Le Parc A, et al. Electrophoresis. 2014 Jun;35(11):1560-70. doi: 10.1002/elps.201300619. Epub 2014 Mar 19. Electrophoresis. 2014. PMID: 24519758 Free PMC article. - The one-pot multienzyme (OPME) synthesis of human blood group H antigens and a human milk oligosaccharide (HMOS) with highly active Thermosynechococcus elongates α1-2-fucosyltransferase.
Zhao C, Wu Y, Yu H, Shah IM, Li Y, Zeng J, Liu B, Mills DA, Chen X. Zhao C, et al. Chem Commun (Camb). 2016 Mar 11;52(20):3899-902. doi: 10.1039/c5cc10646j. Epub 2016 Feb 11. Chem Commun (Camb). 2016. PMID: 26864394 Free PMC article.
References
- Albrecht S, Schols HA, van den Heuvel EGHM, Voragen AGJ, Gruppen H. CE-LIF-MSn profiling of oligosaccharides in human milk and feces of breast-fed babies. Electrophoresis. 2010;31:1264–1273. - PubMed
- Barile D, Marotta M, Chu C, Mehra R, Grimm R, Lebrilla CB, German JB. Neutral and acidic oligosaccharides in Holstein-Friesian colostrum during the first 3 days of lactation measured by high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. J Dairy Sci. 2010;93:3940–3949. - PMC - PubMed
- Barile D, Meyrand M, Lebrilla CB, German JB. Examining bioactive components of milk sources of complex oligosaccharides (part 2) Agro Food Ind Hi Tech. 2011;22:37–39.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources