The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors - PubMed (original) (raw)
The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors
Gijs A Versteeg et al. Immunity. 2013.
Abstract
Innate immunity conferred by the type I interferon is critical for antiviral defense. To date only a limited number of tripartite motif (TRIM) proteins have been implicated in modulation of innate immunity and anti-microbial activity. Here we report the complementary DNA cloning and systematic analysis of all known 75 human TRIMs. We demonstrate that roughly half of the 75 TRIM-family members enhanced the innate immune response and that they do this at multiple levels in signaling pathways. Moreover, messenger RNA levels and localization of most of these TRIMs were found to be altered during viral infection, suggesting that their regulatory activities are highly controlled at both pre- and posttranscriptional levels. Taken together, our data demonstrate a very considerable dedication of this large protein family to the positive regulation of the antiviral response, which supports the notion that this family of proteins evolved as a component of innate immunity.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
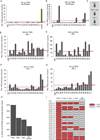
Fig. 1. see also Fig. S1 and Table S1. An unprecedented number of TRIMs enhances innate immune responses
(a) All known TRIM splice variant sequences were mapped using SpliceMiner and the number of distinct proteins they encode determined. These data were used to determine which of previously identified conserved domains they harbor by NCBI CCD. (b) HEK-293T cells were transfected with plasmids encoding individual TRIM proteins. At 48 h p.t. cells were treated with DMSO or lactacystin and subsequently expression of tagged TRIM proteins was analyzed by immunoblot. (c) TRIM21 and TRIM25 were expressed for 50 ng or 500 ng plasmid by transfection in HEK-293T cells in the presence of the limiting amount of 2 ng constitutively active RIG-I(2CARD) plasmid and assayed for their ability to further enhance the IFNβ promoter. Data are represented as mean of triplicates +/− SD. (d) All TRIM proteins were analyzed for their ability to further enhance IFNβ, NF-κB or ISRE promoter activation by a limiting amount of constitutively active RIGI( 2CARD). TRIMs enhancing reporter activity above average were marked as hits and plotted in a heat-map combining the results of all three different promoters in the presence of either 50 ng or 500 ng TRIM plasmid. (e) Overlap and separation of TRIMs enhancing innate immunity in reporter assays at 50 ng and 500 ng plasmid. (f) Distribution of screen hits amongst the TRIM sub-groups as defined by Ozato et al. (Ozato et al., 2008). TRIMs which lack at least one domain in the N-terminal RBCC (RING, B Box, Coiled-coil) domains are shown as unclassified. TRIMs that activate at least one promoter above average are shown in green. TRIM C-terminal domains: ARF, ADP ribosylation factor-like; BR, bromodomain; COS, C-terminal subgroup.
Fig. 2. see also Fig. S2. Most TRIMs are innate immune enhancers and act at different levels of innate immune signaling
HEK-293T cells were transfected with (a) 50 ng or (b) 500 ng TRIM expression plasmid in the absence of additional stimulus and analyzed for ISRE reporter induction. (c) Key regulatory molecules in type I IFN induction used for subsequent assays. To elucidate the level at which individual TRIMs act as enhancers, HEK-293T cells were transfected with individual TRIMs and limiting amounts of (d) TBK1, (e) IKKε and (f and g) constitutively active IRF-3. Data are represented as mean IFNβ reporter induction relative to GST controls +/− SD. (h) The percentage of TRIMs enhancing a particular stimulus were plotted relative to the total number identified as positive hits at 500 ng TRIM plasmid in the presence of RIG-I(2CARD). (i) Heatmap summary of pathway mapping; TRIMs enhancing particular stimuli above average are marked in red.
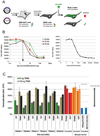
Fig. 3. TRIM proteins enhance the production of antiviral cytokines in a RING-domaindependent manner
(a) Schematic overview of assay. (b) HEK-293T cells were transfected with increasing amounts of RIG-I(2CARD). Supernatants were harvest at 24, 36 and 48 h p.t., which were used to incubate Vero cells for 24 h. As a reference universal IFN dilutions were used (right panel). These Vero cells were infected with VSV-GFP (4 PFU/cell) for 7 h and their GFP expression determined using a plate reader. (c) HEK-293T cells were transfected with a limiting amount of RIGI( 2CARD) plasmid (2 ng) and cotransfected with 50 or 150 ng of wildtype or RING point-mutant TRIM plasmid. At 32 h p.t. supernatants were transferred to Vero cells for 24 h, followed by VSVGFP infection for 7 h. GFP expression was determined using a plate reader. Data are represented as mean relative to the background in mock-infected cells +/− SD.
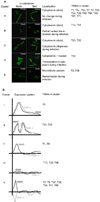
Fig. 4. see also Fig. S3-5. TRIMs are complexly regulated at both the transcriptional and posttranscriptional level
(a) HeLa cells were transfected with tagged TRIM expression plasmids and incubated for 32 h. Subsequently, cells were infected with SeV for 8 h. After fixing, cells were stained using HA and V5 antibodies recognizing the tagged TRIM proteins. The resulting localization data were organized in five distinct clusters by similar localization and relocalization patterns. The depicted immune-fluorescence figures are representative samples for localization patterns in each distinct cluster. (b) Lung-derived A549 and monocyte-derived THP-1 cells were treated with 1000 IU/mL universal IFNβ or infected with SeV. At 3, 6, and 9 h p.t. RNA was isolated and analyzed by real-time RT-PCR for TRIM mRNA expression. The depicted expression figures were drawn by hand to summarize TRIM mRNA expression patterns in each distinct cluster depicted in Fig. S5. TRIM expression regulation was in almost all instances similar upon IFNβ treatment and SeV infection, and thus represented as single lines.
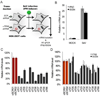
Fig. 5. Stable mRNA targeting of endogenous TRIMs decreases IFNβ and ISG54 upregulation during SeV infection
(a) Schematic overview of assay. HEK-293T cells were transduced with lentiviruses expressing shRNAs targeting individual TRIMs. After puromycin selection, transduced polyclonal cells were infected with SeV and analyzed in triplicate by specific real-time RT-PCR for (b) IFN and ISG54 induction in cells expressing a scrambled shRNA, (c) specific TRIM expression in all mRNA targeted cell pools and (d) IFNβ and ISG54 stimulation by SeV in TRIM mRNA targeted cells. Data are represented as mean +/− SD.
Fig. 6. see also Fig. S6. mRNA targeting of endogenous TRIMs in MDDCs attenuates cytokine expression upon LPS stimulation
MDDCs transduced with lentiviruses expressing TRIM-specific shRNAs were stimulated with LPS. Each condition was repeated in four different donors, with 2-4 wells per condition per donor. (a) At 2 h p.t. cells were harvested and their IFNβ mRNA levels determined by RT-qPCR and plotted as fold induction over mock-induced samples. (b) Similarly, TNFα, IL-6 and IL-8 mRNA levels were determined and plotted as a heat map representing the fraction of donors in which expression of a particular cytokine was >10% affected compared to the scrambled shRNA control. Red-orange colors: 50% or more of the donors exhibited consistent attenuation of the cytokine response upon TRIM mRNA targeting without an equal number of donors with the opposite phenotype. Green colors: 50% or more of the donors consistently upregulated cytokine responses during TRIM mRNA targeting without an equal number of donors with the opposite phenotype. (c) MDDCs from donor 3 and 4 expressing TRIM5 or TRIM38 specific shRNAs, were infected with SeV. At 4 h p.i. their cytokine mRNA levels were determined by RT-qPCR and plotted as fold induction over mock-induced samples. Numbers in parentheses indicate TRIM mRNA targeting levels (higher numbers represent better mRNA targeting); n.d. indicates that mRNA targeting could not be reliably measured resulting from low TRIM mRNA levels. Data are represented as mean +/− SD.
Fig. 7. see also Fig. S7 and Table S2. TRIM proteins differentially regulate various innate signaling pathways
THP1 monocyte-like cells were transduced with lentiviruses expressing a scrambled control shRNA or two individual, different shRNAs targeting the indicated TRIMs. (a) After puromycin selection, mRNA targeting of individual TRIMs was confirmed in polyclonal cell populations by RT-qPCR. Subsequently, these cells were stimulated with (b–c) SeV (3 h; RIG-IMAVS axis), or (d–e) LPS (3 h; TLR4-MyD88-TRIF axis). At the indicated times, total RNA was isolated and IFNβ, ISG54 and IL6 mRNA induction determined by RT-qPCR. Data are represented as mean +/− SD. (f) Heatmap summary of results from cytokine induction in mRNA targeted cells. Half of each oval unit represents each of the two different shRNAs. Samples with ≥ 35% attenuation in cytokine induction are marked in red, or inversely marked green upon ≥ 35% enhancement of cytokine induction. (g) Vero cells were incubated for 24 h with supernatants from THP1 mRNA targeted cells stimulated for 24 h with zymosan. Subsequently, these Vero cells were infected for 7 h with VSV-GFP (4 PFU/cell). Data are represented as mean +/− SD. (h) Model-overview of predicted levels of action of the tested subset of TRIMs.
Similar articles
- TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity.
Rajsbaum R, García-Sastre A, Versteeg GA. Rajsbaum R, et al. J Mol Biol. 2014 Mar 20;426(6):1265-84. doi: 10.1016/j.jmb.2013.12.005. Epub 2013 Dec 12. J Mol Biol. 2014. PMID: 24333484 Free PMC article. Review. - To TRIM the Immunity: From Innate to Adaptive Immunity.
Yang W, Gu Z, Zhang H, Hu H. Yang W, et al. Front Immunol. 2020 Oct 8;11:02157. doi: 10.3389/fimmu.2020.02157. eCollection 2020. Front Immunol. 2020. PMID: 33117334 Free PMC article. - The Roles of TRIMs in Antiviral Innate Immune Signaling.
Shen Z, Wei L, Yu ZB, Yao ZY, Cheng J, Wang YT, Song XT, Li M. Shen Z, et al. Front Cell Infect Microbiol. 2021 Mar 15;11:628275. doi: 10.3389/fcimb.2021.628275. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33791238 Free PMC article. Review. - TRK-Fused Gene (TFG), a protein involved in protein secretion pathways, is an essential component of the antiviral innate immune response.
Khan KA, Marineau A, Doyon P, Acevedo M, Durette É, Gingras AC, Servant MJ. Khan KA, et al. PLoS Pathog. 2021 Jan 7;17(1):e1009111. doi: 10.1371/journal.ppat.1009111. eCollection 2021 Jan. PLoS Pathog. 2021. PMID: 33411856 Free PMC article. - The role of tripartite motif family members in mediating susceptibility to HIV-1 infection.
Rahm N, Telenti A. Rahm N, et al. Curr Opin HIV AIDS. 2012 Mar;7(2):180-6. doi: 10.1097/COH.0b013e32835048e1. Curr Opin HIV AIDS. 2012. PMID: 22258502
Cited by
- TRIM58 Restrains Intestinal Mucosal Inflammation by Negatively Regulating TLR2 in Myeloid Cells.
Eyking A, Ferber F, Köhler S, Reis H, Cario E. Eyking A, et al. J Immunol. 2019 Sep 15;203(6):1636-1649. doi: 10.4049/jimmunol.1900413. Epub 2019 Aug 5. J Immunol. 2019. PMID: 31383741 Free PMC article. - A guide to membrane atg8ylation and autophagy with reflections on immunity.
Deretic V, Lazarou M. Deretic V, et al. J Cell Biol. 2022 Jul 4;221(7):e202203083. doi: 10.1083/jcb.202203083. Epub 2022 Jun 14. J Cell Biol. 2022. PMID: 35699692 Free PMC article. - The Andes Orthohantavirus NSs Protein Antagonizes the Type I Interferon Response by Inhibiting MAVS Signaling.
Vera-Otarola J, Solis L, Lowy F, Olguín V, Angulo J, Pino K, Tischler ND, Otth C, Padula P, López-Lastra M. Vera-Otarola J, et al. J Virol. 2020 Jun 16;94(13):e00454-20. doi: 10.1128/JVI.00454-20. Print 2020 Jun 16. J Virol. 2020. PMID: 32321811 Free PMC article. - Molecular mechanism of influenza A NS1-mediated TRIM25 recognition and inhibition.
Koliopoulos MG, Lethier M, van der Veen AG, Haubrich K, Hennig J, Kowalinski E, Stevens RV, Martin SR, Reis e Sousa C, Cusack S, Rittinger K. Koliopoulos MG, et al. Nat Commun. 2018 May 8;9(1):1820. doi: 10.1038/s41467-018-04214-8. Nat Commun. 2018. PMID: 29739942 Free PMC article. - Genome editing of FTR42 improves zebrafish survival against virus infection by enhancing IFN immunity.
Qu ZL, Gong XY, An LL, Sun HY, Guo WH, Luan HY, Wu MY, Dan C, Gui JF, Zhang YB. Qu ZL, et al. iScience. 2024 Mar 12;27(4):109497. doi: 10.1016/j.isci.2024.109497. eCollection 2024 Apr 19. iScience. 2024. PMID: 38550983 Free PMC article.
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. - PubMed
- Berger G, Durand S, Goujon C, Nguyen XN, Cordeil S, Darlix JL, Cimarelli A. A simple, versatile and efficient method to genetically modify human monocyte-derived dendritic cells with HIV-1-derived lentiviral vectors. Nat Protoc. 2011;6:806–816. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DA033733/DA/NIDA NIH HHS/United States
- P01AI090935/AI/NIAID NIH HHS/United States
- U54 AI57158/AI/NIAID NIH HHS/United States
- U19 AI083025/AI/NIAID NIH HHS/United States
- U54 AI057158/AI/NIAID NIH HHS/United States
- U19AI83025/AI/NIAID NIH HHS/United States
- R01 DA033773/DA/NIDA NIH HHS/United States
- P01 AI090935/AI/NIAID NIH HHS/United States
- R01DA033733/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources