Understanding the role of amphipathic helices in N-BAR domain driven membrane remodeling - PubMed (original) (raw)
Understanding the role of amphipathic helices in N-BAR domain driven membrane remodeling
Haosheng Cui et al. Biophys J. 2013.
Abstract
Endophilin N-BAR (N-terminal helix and Bin/amphiphysin/Rvs) domain tubulates and vesiculates lipid membranes in vitro via its crescent-shaped dimer and four amphipathic helices that penetrate into membranes as wedges. Like F-BAR domains, endophilin N-BAR also forms a scaffold on membrane tubes. Unlike F-BARs, endophilin N-BARs have N-terminal H0 amphipathic helices that are proposed to interact with other N-BARs in oligomer lattices. Recent cryo-electron microscopy reconstructions shed light on the organization of the N-BAR lattice coats on a nanometer scale. However, because of the resolution of the reconstructions, the precise positioning of the amphipathic helices is still ambiguous. In this work, we applied a coarse-grained model to study various membrane remodeling scenarios induced by endophilin N-BARs. We found that H0 helices of N-BARs prefer to align in an antiparallel manner at two ends of the protein to form a stable lattice. The deletion of H0 helices causes disruption of the lattice. In addition, we analyzed the persistence lengths of the protein-coated tubes and found that the stiffness of endophilin N-BAR-coated tubules qualitatively agrees with previous experimental work studying N-BAR-coated tubules. Large-scale simulations on membrane liposomes revealed a systematic relation between H0 helix density and local membrane curvature fluctuations. The data also suggest that the H0 helix is required for BARs to form organized structures on the liposome, further illustrating its important function.
Copyright © 2013 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
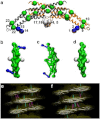
Figure 1
CG model of endophilin N-BARs is shown in panel a. The BAR domain main arch CG sites are colored in green, H0 helices are colored in blue, and the insert helices in white. The sites are assigned based on essential dynamics coarse-graining calculations. Panels b to d show top views of zigzag N-BAR, triad N-BAR, and BAR domain, respectively. Panels e and f show the zigzag model and the triad model fitted into the cryo-EM map, respectively.
Figure 2
CG simulation snapshots of membrane tubes coated by zigzag N-BARs and triad N-BARs. Panels a and b show the initial and final configurations of zigzag simulation and panels c and d show the initial and final configurations of the triad system. The lattice structure remains more organized in the zigzag system.
Figure 3
Order of the N-BAR protein coats. Panel a illustrates the local orientation order parameter, defined by the cosine of the orientational difference between two BARs. Panel b shows a snapshot of the protein lattice, colored from blue to red as the order parameter decreases. Panel c plots the order parameters of the zigzag N-BAR system and the triad N-BAR system as functions of time. The lattice of the triad system is significantly disrupted at the end of the simulation.
Figure 4
Simulation snapshots of BAR domain-coated membrane are shown; panel a is the initial configuration and panel b is the final state of the simulation. Panel c shows the local orientational order parameters calculated for multiple trajectories in the N-BAR system and BAR system. The N-BAR system stays much more organized after the simulation compared to the BAR system with H0 helices deleted.
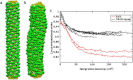
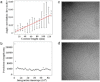
Figure 5
Persistence length calculations. Panel a shows an example of the regression used to compute the persistence length for one membrane tube at one time frame; error bars show the standard deviations of the value of angle correlation for segments that have the same contour lengths. Panel b plots the persistence length for one tube over the simulation as a function of simulation time, demonstrating that the persistence length converges after 50 million CG MD time steps. Panels c and d show representative experimental EM images of endophilin N-BAR-coated tubes.
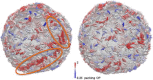
Figure 6
Representative snapshots of the N-BAR system (left) and the BAR-coated liposome systems (right) after 5 million CG MD timestep simulations. The proteins are colored by a packing order parameter that is explained in the main text. Circles in the left panel highlight the region where proteins gather into structures that have similar orientations. This kind of clustering is missing in the BAR domain system in the right panel. The data are consistent with the hypothesis that BAR domains require H0 helices to form arrays and strings on the liposome.
Figure 7
EM image showing aligned arrays of endophilin N-BARs on a liposome surface in an area that is not yet tubulated.
Similar articles
- Roles of amphipathic helices and the bin/amphiphysin/rvs (BAR) domain of endophilin in membrane curvature generation.
Jao CC, Hegde BG, Gallop JL, Hegde PB, McMahon HT, Haworth IS, Langen R. Jao CC, et al. J Biol Chem. 2010 Jun 25;285(26):20164-70. doi: 10.1074/jbc.M110.127811. Epub 2010 Apr 23. J Biol Chem. 2010. PMID: 20418375 Free PMC article. - Structural basis of membrane bending by the N-BAR protein endophilin.
Mim C, Cui H, Gawronski-Salerno JA, Frost A, Lyman E, Voth GA, Unger VM. Mim C, et al. Cell. 2012 Mar 30;149(1):137-45. doi: 10.1016/j.cell.2012.01.048. Cell. 2012. PMID: 22464326 Free PMC article. - The N-Terminal Amphipathic Helix of Endophilin Does Not Contribute to Its Molecular Curvature Generation Capacity.
Chen Z, Zhu C, Kuo CJ, Robustelli J, Baumgart T. Chen Z, et al. J Am Chem Soc. 2016 Nov 9;138(44):14616-14622. doi: 10.1021/jacs.6b06820. Epub 2016 Oct 28. J Am Chem Soc. 2016. PMID: 27755867 Free PMC article. - BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature.
Itoh T, De Camilli P. Itoh T, et al. Biochim Biophys Acta. 2006 Aug;1761(8):897-912. doi: 10.1016/j.bbalip.2006.06.015. Epub 2006 Jul 28. Biochim Biophys Acta. 2006. PMID: 16938488 Review. - A unifying mechanism accounts for sensing of membrane curvature by BAR domains, amphipathic helices and membrane-anchored proteins.
Bhatia VK, Hatzakis NS, Stamou D. Bhatia VK, et al. Semin Cell Dev Biol. 2010 Jun;21(4):381-90. doi: 10.1016/j.semcdb.2009.12.004. Epub 2009 Dec 16. Semin Cell Dev Biol. 2010. PMID: 20006726 Review.
Cited by
- The endophilin curvature-sensitive motif requires electrostatic guidance to recycle synaptic vesicles in vivo.
Zhang L, Wang Y, Dong Y, Pant A, Liu Y, Masserman L, Xu Y, McLaughlin RN Jr, Bai J. Zhang L, et al. Dev Cell. 2022 Mar 28;57(6):750-766.e5. doi: 10.1016/j.devcel.2022.02.021. Epub 2022 Mar 17. Dev Cell. 2022. PMID: 35303431 Free PMC article. - Tubulation by amphiphysin requires concentration-dependent switching from wedging to scaffolding.
Isas JM, Ambroso MR, Hegde PB, Langen J, Langen R. Isas JM, et al. Structure. 2015 May 5;23(5):873-881. doi: 10.1016/j.str.2015.02.014. Epub 2015 Apr 9. Structure. 2015. PMID: 25865245 Free PMC article. - Dynamics and instabilities of lipid bilayer membrane shapes.
Shi Z, Baumgart T. Shi Z, et al. Adv Colloid Interface Sci. 2014 Jun;208:76-88. doi: 10.1016/j.cis.2014.01.004. Epub 2014 Jan 25. Adv Colloid Interface Sci. 2014. PMID: 24529968 Free PMC article. Review. - ACAP1 assembles into an unusual protein lattice for membrane deformation through multiple stages.
Chan C, Pang X, Zhang Y, Niu T, Yang S, Zhao D, Li J, Lu L, Hsu VW, Zhou J, Sun F, Fan J. Chan C, et al. PLoS Comput Biol. 2019 Jul 10;15(7):e1007081. doi: 10.1371/journal.pcbi.1007081. eCollection 2019 Jul. PLoS Comput Biol. 2019. PMID: 31291238 Free PMC article. - Mechanisms of negative membrane curvature sensing and generation by ESCRT III subunit Snf7.
Nepal B, Sepehri A, Lazaridis T. Nepal B, et al. Protein Sci. 2020 Jun;29(6):1473-1485. doi: 10.1002/pro.3851. Epub 2020 Mar 18. Protein Sci. 2020. PMID: 32142182 Free PMC article.
References
- McMahon H.T., Gallop J.L. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. - PubMed
- Slepnev V.I., De Camilli P. Accessory factors in clathrin-dependent synaptic vesicle endocytosis. Nat. Rev. Neurosci. 2000;1:161–172. - PubMed
- Zimmerberg J., Kozlov M.M. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 2006;7:9–19. - PubMed
- Peter B.J., Kent H.M., McMahon H.T. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. - PubMed
- Weissenhorn W. Crystal structure of the endophilin-A1 BAR domain. J. Mol. Biol. 2005;351:653–661. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RR017573/RR/NCRR NIH HHS/United States
- P41 GM103310/GM/NIGMS NIH HHS/United States
- GM094479/GM/NIGMS NIH HHS/United States
- R01 GM063796/GM/NIGMS NIH HHS/United States
- R01 GM094479/GM/NIGMS NIH HHS/United States
- DA24101/DA/NIDA NIH HHS/United States
- P41 RR017573/RR/NCRR NIH HHS/United States
- R21 DA024101/DA/NIDA NIH HHS/United States
- GM063796/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources