The Arabidopsis At1g30680 gene encodes a homologue to the phage T7 gp4 protein that has both DNA primase and DNA helicase activities - PubMed (original) (raw)
The Arabidopsis At1g30680 gene encodes a homologue to the phage T7 gp4 protein that has both DNA primase and DNA helicase activities
Joann Diray-Arce et al. BMC Plant Biol. 2013.
Abstract
Background: The Arabidopsis thaliana genome encodes a homologue of the full-length bacteriophage T7 gp4 protein, which is also homologous to the eukaryotic Twinkle protein. While the phage protein has both DNA primase and DNA helicase activities, in animal cells Twinkle is localized to mitochondria and has only DNA helicase activity due to sequence changes in the DNA primase domain. However, Arabidopsis and other plant Twinkle homologues retain sequence homology for both functional domains of the phage protein. The Arabidopsis Twinkle homologue has been shown by others to be dual targeted to mitochondria and chloroplasts.
Results: To determine the functional activity of the Arabidopsis protein we obtained the gene for the full-length Arabidopsis protein and expressed it in bacteria. The purified protein was shown to have both DNA primase and DNA helicase activities. Western blot and qRT-PCR analysis indicated that the Arabidopsis gene is expressed most abundantly in young leaves and shoot apex tissue, as expected if this protein plays a role in organelle DNA replication. This expression is closely correlated with the expression of organelle-localized DNA polymerase in the same tissues. Homologues from other plant species show close similarity by phylogenetic analysis.
Conclusions: The results presented here indicate that the Arabidopsis phage T7 gp4/Twinkle homologue has both DNA primase and DNA helicase activities and may provide these functions for organelle DNA replication.
Figures
Figure 1
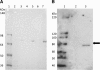
Purification of the recombinant protein. Panel A shows the Coomassie Blue-stained gel, with increasing amounts of the purified recombinant (lanes 3, 5 and 7) and control (lanes 2, 4 and 6) protein, from left to right. Lane 1, protein molecular weight markers (Invitrogen SeeBlue 2 markers). Lanes 2 and 3, 0.195 ng; lanes 4 and 5, 0.39 ng; lanes 6 and 7, 0.585 ng. Panel B shows a western blot of the purified protein using antibody against the Arabidopsis Twinkle homologue. Lane 1 contains molecular weight markers (Invitrogen Magic Markers). Lane 2, control protein; lane 3, 0.5 ng purified recombinant protein. The arrow at the right indicates 80 kDa, the length of the full-length Arabidopsis gene product. The recombinant protein is slightly smaller (~74 kDa) as it lacks the N-terminal localization sequence.
Figure 2
DNA primase assay. The recombinant Twinkle homologue purified from E. coli cells was tested for DNA primase activity. Panel A, lane L (DNA single-base ladder), oligo dT9-18 included as size markers (same for panel B). Lane T, reaction products with the recombinant protein. Lane C, reaction products using a bacterial fraction with the empty vector as control. Panel B shows incorporation of primers into high molecular weight DNA in the presence (lane 6) but not the absence (lane 5) of E. coli DNA polymerase I and dNTPs. Lanes 3 and 4 are the control protein fraction in the absence (lane 3) and presence (lane 4) of DNA polymerase I and dNTPs.
Figure 3
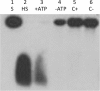
DNA helicase assay. The recombinant Twinkle homologue purified from E. coli cells was tested for DNA helicase activity as described in the text. Lane 1 is the control substrate (S). Lane 2 is the heated control (HS), showing separation of the short labeled oligo from the substrate, which runs in this gel as a leading band with a diffuse smear; lane 3 (T+ATP), reaction using the purified recombinant protein with ATP; lane 4 (T-ATP), same reaction without ATP, lane 5 (C+ATP), control protein from E. coli cells lacking the expression construct with ATP, lane 6 (C–ATP), same reaction but without ATP.
Figure 4
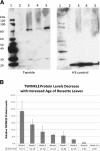
Western blot analysis of Arabidopsis Twinkle homologue expression. A. Lane 1, Molecular weight markers, Lane 2, leaf tissue from 6-week plants; lane 3, shoot apex tissue; lane 4, total plant tissue protein; lane 5, cotyledon protein. The panel on the left was incubated with antibody against the Twinkle protein. The panel on the right was incubated with histone H3 antibody as a loading control. B. Relative levels of Twinkle protein relative to a nuclear tubulin protein control in Arabidopsis seedlings harvested at the times indicated. The average of three independent western blots is shown for each time point (weeks 1–5 and 10). Error bars indicate the SEM (standard error of the mean).
Figure 5
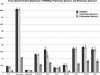
RT-qPCR analysis of the Arabidopsis Twinkle homologue gene expression relative to organellar localized DNA polymerases in various tissues. The relative abundance of Twinkle and the two organellar DNA polymerases (Polymerase gamma I and Polymerase gamma II) is shown. Expression varied among selected organs with highest expression in the shoot apex. The relative expression of Twinkle follows the expression levels of DNA polymerase gamma I. Error bars indicate SEM of three replicates. The Y axis indicates relative expression (log2) normalized to nuclear actin gene expression. Inflor, inflorescence.
Figure 6
A. DNA sequence alignments of some Twinkle primase domain conserved regions to show the extent of changes between different organisms. The DNA sequences for the Twinkle protein from T7, Arabidopsis (At1g30680), human (Hs) and Drosophila (Dm) are shown for the conserved motifs I, IV and V. The locations of the cysteine residues in Motif I are indicated above the sequence while the corresponding codon sequence is underlined in the DNA sequence. The central conserved elements of each motif are shaded yellow. Base differences from the T7 gp4 sequence are shaded dark blue with white lettering.
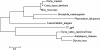
Figure 7
Phylogenetic analysis of the T7 gp4 protein, plant homologues, and selected eukaryotic Twinkle protein homologues. Molecular phylogenetic analysis was performed using the maximum likelihood method. The scale bar indicates the number of substitutions per site.
Similar articles
- Chimeric proteins constructed from bacteriophage T7 gp4 and a putative primase-helicase from Arabidopsis thaliana.
Towle-Weicksel JB, Cao Y, Crislip LJ, Thurlow DL, Crampton DJ. Towle-Weicksel JB, et al. Mol Biol Rep. 2014 Dec;41(12):7783-95. doi: 10.1007/s11033-014-3671-y. Epub 2014 Aug 7. Mol Biol Rep. 2014. PMID: 25098604 - The plant organellar primase-helicase directs template recognition and primosome assembly via its zinc finger domain.
Peralta-Castro A, Cordoba-Andrade F, Díaz-Quezada C, Sotelo-Mundo R, Winkler R, Brieba LG. Peralta-Castro A, et al. BMC Plant Biol. 2023 Oct 6;23(1):467. doi: 10.1186/s12870-023-04477-4. BMC Plant Biol. 2023. PMID: 37803262 Free PMC article. - Heterohexamer of 56- and 63-kDa Gene 4 Helicase-Primase of Bacteriophage T7 in DNA Replication.
Zhang H, Lee SJ, Kulczyk AW, Zhu B, Richardson CC. Zhang H, et al. J Biol Chem. 2012 Oct 5;287(41):34273-87. doi: 10.1074/jbc.M112.401158. Epub 2012 Aug 10. J Biol Chem. 2012. PMID: 22887996 Free PMC article. - Mechanisms of a ring shaped helicase.
Donmez I, Patel SS. Donmez I, et al. Nucleic Acids Res. 2006;34(15):4216-24. doi: 10.1093/nar/gkl508. Epub 2006 Aug 25. Nucleic Acids Res. 2006. PMID: 16935879 Free PMC article. Review. - Crawling and wiggling on DNA: structural insights to the mechanism of DNA unwinding by helicases.
Marians KJ. Marians KJ. Structure. 2000 Dec 15;8(12):R227-35. doi: 10.1016/s0969-2126(00)00539-6. Structure. 2000. PMID: 11188698 Review.
Cited by
- Primase promotes the competition between transcription and replication on the same template strand resulting in DNA damage.
Zhang W, Yang Z, Wang W, Sun Q. Zhang W, et al. Nat Commun. 2024 Jan 2;15(1):73. doi: 10.1038/s41467-023-44443-0. Nat Commun. 2024. PMID: 38168108 Free PMC article. - Isolation and characterization of the Staphylococcus aureus bacteriophage vB_SauS_SA2.
Wang J, Zhao F, Sun H, Wang Q, Zhang C, Liu W, Zou L, Pan Q, Ren H. Wang J, et al. AIMS Microbiol. 2019 Sep 27;5(3):285-307. doi: 10.3934/microbiol.2019.3.285. eCollection 2019. AIMS Microbiol. 2019. PMID: 31663062 Free PMC article. - Mitochondrial DNA replication: a PrimPol perspective.
Bailey LJ, Doherty AJ. Bailey LJ, et al. Biochem Soc Trans. 2017 Apr 15;45(2):513-529. doi: 10.1042/BST20160162. Biochem Soc Trans. 2017. PMID: 28408491 Free PMC article. Review. - Minireview: DNA replication in plant mitochondria.
Cupp JD, Nielsen BL. Cupp JD, et al. Mitochondrion. 2014 Nov;19 Pt B:231-7. doi: 10.1016/j.mito.2014.03.008. Epub 2014 Mar 26. Mitochondrion. 2014. PMID: 24681310 Free PMC article. Review. - Chimeric proteins constructed from bacteriophage T7 gp4 and a putative primase-helicase from Arabidopsis thaliana.
Towle-Weicksel JB, Cao Y, Crislip LJ, Thurlow DL, Crampton DJ. Towle-Weicksel JB, et al. Mol Biol Rep. 2014 Dec;41(12):7783-95. doi: 10.1007/s11033-014-3671-y. Epub 2014 Aug 7. Mol Biol Rep. 2014. PMID: 25098604
References
- Kornberg A, Baker T. DNA Replication, vol 2. New York, New York: W.H. Freeman and Co.; 1991.
- Tougu K, Peng H, Marians KJ. Identification of a domain of Escherichia coli primase required for functional interaction with the DnaB helicase at the replication fork. J Biol Chem. 1994;269(6):4675–4682. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous