Jmjd3 inhibits reprogramming by upregulating expression of INK4a/Arf and targeting PHF20 for ubiquitination - PubMed (original) (raw)
Jmjd3 inhibits reprogramming by upregulating expression of INK4a/Arf and targeting PHF20 for ubiquitination
Wei Zhao et al. Cell. 2013.
Abstract
Although somatic cell reprogramming to generate inducible pluripotent stem cells (iPSCs) is associated with profound epigenetic changes, the roles and mechanisms of epigenetic factors in this process remain poorly understood. Here, we identify Jmjd3 as a potent negative regulator of reprogramming. Jmjd3-deficient MEFs produced significantly more iPSC colonies than did wild-type cells, whereas ectopic expression of Jmjd3 markedly inhibited reprogramming. We show that the inhibitory effects of Jmjd3 are produced through both histone demethylase-dependent and -independent pathways. The latter pathway involves Jmjd3 targeting of PHF20 for ubiquitination and degradation via recruitment of an E3 ligase, Trim26. Importantly, PHF20-deficient MEFs could not be converted to fully reprogrammed iPSCs, even with knockdown of Jmjd3, Ink4a, or p21, indicating that PHF20 is required for reprogramming. Our findings demonstrate, to the best of our knowledge, a previously unrecognized role of Jmjd3 in cellular reprogramming and provide molecular insight into the mechanisms by which the Jmjd3-PHF20 axis controls this process.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
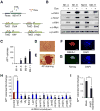
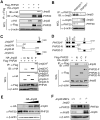
Figure 1. Identification of Jmjd3 and Other Key Epigenetic Factors that Regulate Reprogramming
(A) Outline of generation of transgenic mice expressing rtTA, together with Oct4 (O), Sox2 (S), Klf4 (K) and Myc (M) (OSKM, 4F) under control of a tetracycline-on promoter (Tet-O). (B) Western blot analysis of 4F expression in Tet-O MEFs treated with or without Dox. (C) Alkaline phosphatase (AP)-positive colonies were counted at day12 after Dox treatment. (D) Bright field images of an iPSC colony derived from Tet-O 4F MEFs. (E-G) Staining of representative iPSC colonies with antibodies against AP, stage-specific embryonic antigen 1 (SSEA1) and Nanog. Scale bars in panels D, E, F and G, 50μm (H) Fold changes in number of AP-positive colonies generated from Tet-O 4F MEFs transduced with specific shRNA, compared with control shRNA. AP-positive colonies were counted on day14 after Dox treatment. (I) Fold changes in number of AP-positive colonies generated from Tet-O 4F MEFs transduced with Jmjd3 expression or empty vector. Ectopic expression of Jmjd3 inhibits reprogramming. The data in panels H and I are reported as the means ± SD with indicated significance (*p <0.05, **p < 0.01 ***p < 0.001 by Student's t test). See also Figure S1.
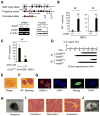
Figure 2. Jmjd3 Ablation Enhances the Efficiency and Kinetics of Reprogramming
(A) Establishment of Jmjd3 knockout mice and _Jmjd3_-deficient MEFs. PCR and immunoblot (IB) analyses confirmed loss of Jmjd3 expression in _Jmjd3_-deficient MEFs. (B) AP-positive colonies were counted on day 14 of 4F-mediated or on day 21 of 3F-mediated reprogramming of WT and _Jmjd3_-deficient MEF cells. (C) AP-positive colonies were counted on day 14 of 4F-mediated reprogramming after tamoxifen treatment of Ezh2f/f :Cre-ESR1 MEFs. (D) Schematic representation of the time of AP+ colony appearance during reprogramming. (E) Bright-field image of a representative iPSC colony generated from _Jmjd3_-deficient MEFs. (F and G) Staining of representative iPSC colonies with antibodies against AP, SSEA1 and Nanog. Scale bars, 50 μm. (H and I) Photomicrograph and HE (hematoxylin and eosin) staining of teratomas generated from immune-deficient mice harboring a _Jmjd_3-deficient iPSC clone. Three layers of teratoma (ectoderm, mesoderm and endoderm) are presented. Scale bars, 100 μm. (J) _Jmjd3_−/− MEF-derived iPSCs were pluripotent and contributed to chimeras after injection into BALB/C host blastocysts as indicated by coat color. The data in panels B and C are reported as means ± SD with indicated significance (*p < 0.05, **p < 0.01 by Student's t test).
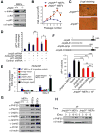
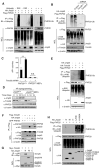
Figure 3. Identification of Jmjd3 Targets Responsible for Enhanced Reprogramming
(A) Western blot analysis of Ink4a, Arf and p21 in WT and _Jmjd3_-deficient MEFs at passage 4. (B) Growth of WT versus _Jmjd3_-deficient MEFs. (C) β-gal staining of WT versus _Jmjd3_-deficient MEFs at passage 4. (D) Number of AP-positive iPSC colonies counted on day 14 of 4F-mediated reprogramming of WT MEFs transduced with _Jmjd3_-, _Ink4a/Arf_-, _p21_-specific or control shRNAs. (E) _Jmjd3_-deficient MEFs transduced with lentiviruses expressing Jmjd3 or its mutants together with 4F for 3 days. Expression of Ink4a/Arf was determined by real-time PCR analysis and was normalized by internal control beta-actin. (F) Number of AP-positive iPSC colonies counted on day 11 of 4F-mediated reprogramming of _Jmjd3_-deficient MEFs transduced with lentiviruses expressing Jmjd3 or its mutants. (G) Western blot analysis of 7 epigenetic proteins in WT and _Jmjd3_-deficient MEFs, iPSCs and ES cells. (H) Western blot analysis of PHF20 expression in WT and _Jmjd3_-deficient MEFs during 4F-mediated reprogramming. The data in panels B, D-F are reported as means ± SD with indicated significance (*p < 0.05, **p < 0.01, ***p<0.001). See also Figures S2 and S3.
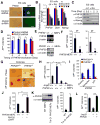
Figure 4. PHF20 Is Essential for Maintenance and Reprogramming of iPSCs
(A) Bright-field and GFP images of ESCs transduced with PHF20-specific or control shRNA. Scale bars, 50 μm. (B) Real-time PCR analysis of PHF20, Oct4 and Nanog expression in ESCs treated with 1 mM RA and LIF withdrawal. (C) Western blot analysis of PHF20, Oct4 and Nanog expression after treatment of ESCs with 1 μM RA and LIF withdrawal. (D) Colony formation after PHF20 knockdown by _PHF20_-specific or a control lentivirus-based constitutive shRNA at different time points (i.e., day 0, 4, 8 or 12) during reprogramming of Tet-O-4F MEFs. AP-positive colonies were counted on day 14. (E) PCR and Western blot analyses of PHF20 expression in WT and _PHF20_-deficient MEFs. (F) AP-positive colony numbers were counted on day 14 of 3F- or 4F-mediated reprogramming of WT and _PHF20_-deficient MEFs. (G) Bright-field images and AP staining of iPSC-like colonies from WT and _PHF20_-deficient MEFs. Scale bars, 50 μm. (H) Number of AP-positive colonies counted on day 14 of 4F-mediated reprogramming of WT and PHF20 single, or Jmjd3 and _PHF20_-double KO MEFs. (I) Number of AP-positive colonies on day 14 of 4F-mediated reprogramming of WT and PHF20 KO MEFs transduced with _Ink4a/Arf_- or p21_-specific or control shRNA. (J) Number of AP-positive colonies counted on day 14 of 4F-mediated reprogramming of WT and PHF20 knockout MEFs with or without PHF20 cDNA expression. (K) Western analysis of PHF20 expression in WT and Tet-O-PHF20 transgenic MEFs expressing rtTA in the presence of Dox. AP-positive colonies were counted on day 14 of 4F-mediated reprogramming of rtTA_-expressing Tet-O-PHF20 transgenic MEFs. (L) Number of AP-positive colonies counted on day 14 of 4F-mediated reprogramming of WT MEFs transduced with Tet-O_-Jmjd3 or Tet-O_-PHF20 in the presence of Dox until day 10. The data in panels D, F and H-L are plotted as means ± SD with indicated significance (*p < 0.05, **p < 0.01). See also Figure S4.
Figure 5. Jmjd3 Interacts with PHF20 and Causes its Degradation
(A) 293T cells were transfected with Flag-PHF20 and HA-Jmjd3. Cell extracts were immunoprecipitated with anti-Flag beads, followed by immunoblot (IB) with an anti-HA antibody. (B) Detection of interaction between Jmjd3 and PHF20 in WT and PHF20 knockout MEFs by immunoprecipitation (IP) with a PHF20 antibody, followed by IB with an anti-Jmjd3 antibody. (C and D) 293T cells were transfected with different HA-Jmjd3 domain constructs (C) and different Flag-PHF20 constructs (D). Cell extracts were immunoprecipitated with anti-Flag beads, followed by IB with an anti-HA antibody. (E) 293T cells were transfected with increasing amounts of HA-Jmjd3 cDNA, followed by IB with anti-PHF20 and anti-HA antibodies. (F) Jmjd3 WT and KO MEF cells were infected with or without Flag-tagged Jmjd3 retrovirus for24 h, and cell extracts were analyzed by IB with anti-PHF20 and anti-Jmjd3 antibodies. See also Figure S5.
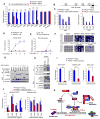
Figure 6. Jmjd3 Targets PHF20 for Ubiquitination by Recruiting an E3 Ligase Trim26
(A) 293T cells transfected with HA-tagged ubiquitin, Flag-tagged PHF20 and HA-tagged Jmjd3. Cell lysates were immunoprecipitated with anti-Flag beads, followed by IB with an anti-ubiquitin antibody. (B) 293T cells transfected with Flag-tagged PHF20, HA-tagged Jmjd3 and different Trim26 shRNA constructs as indicated. Cell lysates were immunoprecipitated with anti-Flag beads and analyzed with an anti-K48 ubiquitin. (C) Number of AP-positive colonies counted on day 14 of 4F-mediated reprogramming of WT and _PHF20_-deficient MEFs transduced with _Trim26_-specific or control shRNAs. Asterisks indicate significant differences between groups (*p < 0.05, **p < 0.01, N.S. p > 0.05). (D) Western blot analysis of Jmjd3, Trim26 and PHF20 expression in WT and _Jmjd3_-deficient MEFs during cellular reprogramming. (E) 293T cells transfected with HA-Trim26, Flag-PHF20 and HA-Jmjd3 as indicated. Cell lysates were immunoprecipitated with anti-Flag beads, followed by IB with an anti-K48 ubiquitin antibody. (F) 293T cells transfected with Flag-Trim26, GFP-PHF20 and HA-Jmjd3 as indicated. Cell lysates were immunoprecipitated with anti-Flag beads, followed by IB with PHF20 and HA antibodies. (G) 293T cells transfected with Myc-Trim26 and HA-Jmjd3 constructs. Cell lysates were immunoprecipitated with anti-HA beads, followed by IB with an anti-Myc antibody. (H) 293T cells transfected with Flag-PHF20 and HA-Jmjd3 constructs. Cell lysates were immunoprecipitated with anti-Flag beads, followed by IB with an anti-K48 ubiquitin antibody. See also Figure S6.
Figure 7. PHF20 is Required for_Oct4_Expression during Reprogramming by Interacting with Wdr5
(A) WT and PHF20 knockout MEFs were reprogrammed by 4F in the presence of Dox for 10 days, followed by Dox withdrawal. Real-time PCR analysis of endogenous Oct4, Sox2, Nanog and other ES genes was performed on day 14. (B) Schematic diagram of Oct4 promoter regions. (C) Determination of PHF20 binding to the CR regions of Oct4 promoter in iPSCs and ESCs by ChIP-qPCR analysis with PHF20-specific antibody. Values are reported as fold enrichment relative to input DNA. (D) ChIP-PCR analysis of timing of PHF20 binding to the Oct4 promoter region during 4F-mediated reprogramming using a PHF20-specific antibody. (E) Real-time PCR analysis of Oct4 expression in rtTA_-expressing WT and Tet-O-PHF20 transgenic MEFs during reprogramming. (F) Bisulfite sequencing of the Oct4 promoter region in ESCs, PHF20+/+ MEFs, PHF20−/− MEFs,iPSCs from PHF20+/+ MEFs, iPS-like (incompletely reprogrammed) cells from PHF20−/− MEFs,and iPSCs from PHF20-expressing PHF20−/− MEFs. Open circles (○) stand for unmethylated CpG dinucleotides, while closed circles (•) for methylated CpGs. (G) 293T cells were transfected with GFP-PHF20, Flag-tagged Wdr5, MLL3, Dpy-30, Ash2L or_Rbbp5. Cell extracts were immunoprecipiated with anti-Flag beads, followed by IB with an anti-PHF20 antibody. (H) Detection of endogenous interaction between PHF20 and Wdr5 complex in iPSCs by IP with a PHF20 antibody, followed by IB with Wdr5, RbBP5, MOF and Ash2L antibodies. (I) ChIP-qPCR analysis of Wdr5, RbBP5 and MOF binding to Oct4 promoter region during 4F-mediated reprogramming of WT and _PHF20_-deficient MEFs. (J) ChIP-qPCR analysis of H3K4me3 and H4K16ac mark of the Oct4 promoter in WT MEF-derived iPSCs and _PHF20_-deficient derived iPSC-like (incompletely reprogrammed) cells. (K) Proposed a working model by which Jmjd3 regulates somatic cell reprogramming. The data in panels A, C-E, I and J are plotted as means ± SD with indicated significance (*p < 0.05, **p < 0.01, by Student's t test). See also Figure S7.
Similar articles
- JMJD3 acts in tandem with KLF4 to facilitate reprogramming to pluripotency.
Huang Y, Zhang H, Wang L, Tang C, Qin X, Wu X, Pan M, Tang Y, Yang Z, Babarinde IA, Lin R, Ji G, Lai Y, Xu X, Su J, Wen X, Satoh T, Ahmed T, Malik V, Ward C, Volpe G, Guo L, Chen J, Sun L, Li Y, Huang X, Bao X, Gao F, Liu B, Zheng H, Jauch R, Lai L, Pan G, Chen J, Testa G, Akira S, Hu J, Pei D, Hutchins AP, Esteban MA, Qin B. Huang Y, et al. Nat Commun. 2020 Oct 8;11(1):5061. doi: 10.1038/s41467-020-18900-z. Nat Commun. 2020. PMID: 33033262 Free PMC article. - JMJD3 suppresses stem cell-like characteristics in breast cancer cells by downregulation of Oct4 independently of its demethylase activity.
Xun J, Wang D, Shen L, Gong J, Gao R, Du L, Chang A, Song X, Xiang R, Tan X. Xun J, et al. Oncotarget. 2017 Mar 28;8(13):21918-21929. doi: 10.18632/oncotarget.15747. Oncotarget. 2017. PMID: 28423536 Free PMC article. - Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS.
Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenführ M, Maertens G, Banck M, Zhou MM, Walsh MJ, Peters G, Gil J. Barradas M, et al. Genes Dev. 2009 May 15;23(10):1177-82. doi: 10.1101/gad.511109. Genes Dev. 2009. PMID: 19451218 Free PMC article. - JMJD3: a critical epigenetic regulator in stem cell fate.
Ding Y, Yao Y, Gong X, Zhuo Q, Chen J, Tian M, Farzaneh M. Ding Y, et al. Cell Commun Signal. 2021 Jul 3;19(1):72. doi: 10.1186/s12964-021-00753-8. Cell Commun Signal. 2021. PMID: 34217316 Free PMC article. Review. - Histone demethylase Jumonji D3 (JMJD3/KDM6B) at the nexus of epigenetic regulation of inflammation and the aging process.
Salminen A, Kaarniranta K, Hiltunen M, Kauppinen A. Salminen A, et al. J Mol Med (Berl). 2014 Oct;92(10):1035-43. doi: 10.1007/s00109-014-1182-x. Epub 2014 Jun 14. J Mol Med (Berl). 2014. PMID: 24925089 Review.
Cited by
- The localization of histone H3K27me3 demethylase Jmjd3 is dynamically regulated.
Kamikawa YF, Donohoe ME. Kamikawa YF, et al. Epigenetics. 2014 Jun;9(6):834-41. doi: 10.4161/epi.28524. Epub 2014 Mar 19. Epigenetics. 2014. PMID: 24646476 Free PMC article. - Epigenetic regulation of T helper cells and intestinal pathogenicity.
Hagihara Y, Yoshimatsu Y, Mikami Y, Takada Y, Mizuno S, Kanai T. Hagihara Y, et al. Semin Immunopathol. 2019 May;41(3):379-399. doi: 10.1007/s00281-019-00732-9. Epub 2019 Mar 19. Semin Immunopathol. 2019. PMID: 30891628 Review. - Jmjd3-mediated epigenetic regulation of inflammatory cytokine gene expression in serum amyloid A-stimulated macrophages.
Yan Q, Sun L, Zhu Z, Wang L, Li S, Ye RD. Yan Q, et al. Cell Signal. 2014 Sep;26(9):1783-91. doi: 10.1016/j.cellsig.2014.03.025. Epub 2014 Apr 1. Cell Signal. 2014. PMID: 24703936 Free PMC article. - Wound healing, cellular regeneration and plasticity: the elegans way.
Vibert L, Daulny A, Jarriault S. Vibert L, et al. Int J Dev Biol. 2018;62(6-7-8):491-505. doi: 10.1387/ijdb.180123sj. Int J Dev Biol. 2018. PMID: 29938761 Free PMC article. Review. - MAP2K6 remodels chromatin and facilitates reprogramming by activating Gatad2b-phosphorylation dependent heterochromatin loosening.
Xing G, Liu Z, Huang L, Zhao D, Wang T, Yuan H, Wu Y, Li L, Long Q, Zhou Y, Hao Z, Liu Y, Lu J, Li S, Zhu J, Wang B, Wang J, Liu J, Chen J, Pei D, Liu X, Chen K. Xing G, et al. Cell Death Differ. 2022 May;29(5):1042-1054. doi: 10.1038/s41418-021-00902-z. Epub 2021 Nov 24. Cell Death Differ. 2022. PMID: 34815549 Free PMC article.
References
- Adams-Cioaba MA, Li Z, Tempel W, Guo Y, Bian C, Li Y, Lam R, Min J. Crystal structures of the Tudor domains of human PHF20 reveal novel structural variations on the Royal Family of proteins. FEBS Lett. 2012;586:859–865. - PubMed
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01CA121191/CA/NCI NIH HHS/United States
- R01CA09327/CA/NCI NIH HHS/United States
- R01 CA116408/CA/NCI NIH HHS/United States
- R01 CA121191/CA/NCI NIH HHS/United States
- R01 DA030338/DA/NIDA NIH HHS/United States
- R01 CA090327/CA/NCI NIH HHS/United States
- R01 CA101795/CA/NCI NIH HHS/United States
- R01CA101795/CA/NCI NIH HHS/United States
- P01 CA094237/CA/NCI NIH HHS/United States
- R01CA116408/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases