The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway - PubMed (original) (raw)
The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway
Lucille C Rankin et al. Nat Immunol. 2013 Apr.
Erratum in
- Nat Immunol. 2013 Apr;14(4):395
- Nat Immunol. 2013 Aug;14(8):877
Abstract
NKp46+ innate lymphoid cells (ILCs) serve important roles in regulating the intestinal microbiota and defense against pathogens. Whether NKp46+ ILCs arise directly from lymphoid tissue-inducer (LTi) cells or represent a separate lineage remains controversial. We report here that the transcription factor T-bet (encoded by Tbx21) was essential for the development of NKp46+ ILCs but not of LTi cells or nuocytes. Deficiency in interleukin 22 (IL-22)-producing NKp46+ ILCs resulted in greater susceptibility of Tbx21-/- mice to intestinal infection. Haploinsufficient T-bet expression resulted in lower expression of the signaling molecule Notch, and Notch signaling was necessary for the transition of LTi cells into NKp46+ ILCs. Furthermore, NKp46+ ILCs differentiated solely from the CD4- LTi population, not the CD4+ LTi population. Our results pinpoint the regulation of Notch signaling by T-bet as a distinct molecular pathway that guides the development of NKp46+ ILCs.
Figures
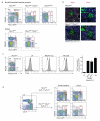
Figure 1. NKp46+ ILCs fail to form in the absence of the transcription factor T-bet
Innate lymphocytes isolated from the small intestinal lamina propria were analysed for in T-bet deficient mice and for T-bet expression. a, Lamina propria cells were isolated from the small intestine (upper panels) and colon (lower panels) of Rorc(γt)gfp/+, Rorc(γt)gfp/+Tbx21−/−, and Rorc(γt)gfp/gfp mice. It should be noted that in the colon, only a single population of NKp46+Rorγt+ cells exists in Tbx21+/+ mice and this population was also lacking in Tbx21−/− mice. Numbers indicate the percentage of live CD45+lin− (CD11c−Gr-1−CD19−CD3ε−CD45+) cells for gated type of cell. Plots are representative of at least 3 independent experiments with three to four mice of each genotype per experiment. b, Small intestine sections from naïve Rorc(γt)gfp/+(upper panels) and Rorc(γt)gfp/+Tbx21−/− (lower panels) mice showing cryptopatches stained with anti-NKp46 (light blue), anti-CD3ε or anti-B220 (red), anti-CD326 (EpcCAM, white) to delineate gut epithelial cells, and endogenous expression of Rorγt-GFP (green). Inset shows NKp46+ staining in Rorγt+ cells. Scale bar, 50μM. Data are representative of at least three independent experiments (_n_=6 per genotype). c, Innate lymphocytes from small intestinal lamina propria expressing different levels of Rorγt-GFP (left panel, gating strategy shown in dot plot) were stained for surface lineage markers, then fixed and permeabilized prior to staining to detect intracellular T-bet (right panels, histograms). Grey histrogram shows T-bet specific stain, dotted line indicates control stain. c, Gating stategy and analyses of individual innate lymphocyte subsets purified by flow cytometry. Tbx21 mRNA was analysed by quantitative PCR and show mean ± s.e.m. normalised to Hprt of triplicate samples showing one representative experiment of three. Dot plots are gated on lin−CD45+ cells. d, Intrinsic T-bet expression is required for NKp46+ ILC development. Mixed foetal liver chimeric mice were generated by transplanting Ly5.1/5.2+ Rorc(γt)gfp/+ and Ly5.2+ Rorc(γt)gfp/+Tbx21−/− cells into lethally irradiated Ly5.1 mice. After 8 weeks, innate lymphocytes were isolated from the lamina propria of the small intestine and colon and the capacity for Tbx21+/+ (Ly5.1+/5.2+) or Tbx21−/− (Ly5.2+) foetal liver cells to reconstitute innate lymphocytes was evaluated. Numbers indicate the frequency of each ILC population within the lin−CD45+ live SYTOX Blue− cell gate which were then examined for expression of α4β7, c-Kit, IL-7R and RORγt-GFP. Data show representative profiles from two independent experiments (_n_=8) with similar results. As gated cells in (c) are fixed, some change in Rorγt-GFP resolution is observed when compared to unfixed cells shown in (a, d).
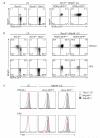
Figure 2. Notch and T-bet regulate the developmental pathway of innate lymphocyte populations
a, In vivo differentiation of ILC subsets in the presence or absence of T-bet. 0.5-1 × 105 LTi cells or NKp46+ ILCs were purified from the small intestinal lamina propria of either _Rorc(γt)gfp/+_or Rorc(γt)gfp/+Tbx21−/− mice then adoptively transferred into Rag2γc-deficient mice. After 14 days, donor lymphocytes were isolated from the small intestinal lamina propria of recipient mice and analysed for the differentiation of innate lymphocyte populations. Data show representative profiles of one of four independent experiments with similar results. Dot plots are gated on lin−CD45+ live donor cells. b, In vitro differentiation of LTi or NKp46+ mature ILCs purified from the small intestinal lamina propria of _Rorc(γt)gfp/+_and Rorc(γt)gfp/+ Tbx21−/− mice cultured on OP9 or OP9-DL1 stromal cells in the presence of SCF, IL-7 and Flt3L and analysed on day 9 of culture. Data are representative of at least 2-4 independent experiments with similar results. c, T-bet expression in ILC subsets differentiated as in b.
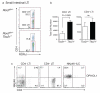
Figure 3. CD4− and not CD4+ LTi cells give rise to NKp46+ ILCs
a. Frequency of CD4− and CD4+ LTi subsets isolated from the small intestine of Tbx21+/+ and _Tbx21_−/− mice. b, Total cell number of CD4− and CD4+ LTi cells in the small intestine of Tbx21+/+ compared with T-bet deficient mice. Data show mean ± s.e.m (n=4/group). c, CD4− LTi, CD4+LTi and NKp46+ ILCs cells were sorted from Tbx21+/+ mice and cultured on OP9-DL1 cells in the presence of IL7, SCF and Flt-3L. Differentiated cells were analyzed for NKp46 expression after 5 days. Data are representative of two independent experiments with similar results.
Figure 4. Notch and T-bet regulate the developmental pathway of innate lymphocyte populations
a, Quantitative RT-PCR analyses of Notch1 and Notch2 in ILCs isolated from the small intestine of mice of the indicated genotypes. RT-PCR analyses show the mean ± s.e.m. of samples analysed in triplicate; ILC subsets show mean percent CD45+lin− (CD11c−Gr-1−CD19−CD3ε−CD45+) PI− cells ± s.e.m. isolated from small intestinal lamina propria. Data are representative of at least three similar experiments. b. Binding of T-bet within the Notch1 and Notch2 genes was analysed using ChIP. Data are the mean enrichment calculated as the percentage enrichment compared to non-immunoprecipitated input DNA ± s.d. from 3 independent samples. c. Rescue of NKp46+ ILC development in LTi cells by retroviral expression of either T-bet or Notch ICS in Tbx21−/− LTi cells and coculture on OP9 or OP9-DL1 stromal cells for 5 days. Shown are flow cytometry plots gated on live Cherry+ cells. Numbers indicate the proportion of cells within the transduced population (Cherry+) that differentiated into NKp46+ ILCs.
Figure 5. CD4−LTi cells, not CD4+LTi cells, give rise to NKp46+ ILCs
(a) Flow cytometry of LTi subsets isolated from the small intestine of Rorc(γt)+/GFP_Tbx21_+/+ and Rorc(γt)+/GFP_Tbx21_−/− mice. Numbers adjacent to outlined areas indicate percent CD4−LTi cells (bottom) and CD4+LTi cells (bottom). (b) Total CD4− and CD4+LTi cells in the small intestine of Rorc(γt)+/GFP_Tbx21_+/+ and Rorc(γt)+/GFP_Tbx21_−/− mice (n = 4 per genotype). NS, not significant; *P = 0.0083 (unpaired Student’s _t_-test). (c) NKp46 expression by CD4−LTi cells, CD4+LTi cells and NKp46+ ILCs sorted from wild-type mice and cultured for 5 d on OP9-DL1 cells in the presence of IL-7, SCF and Flt-3L. Data are representative of at least two experiments (mean and s.e.m. in b).
Similar articles
- TCF-1 controls ILC2 and NKp46+RORγt+ innate lymphocyte differentiation and protection in intestinal inflammation.
Mielke LA, Groom JR, Rankin LC, Seillet C, Masson F, Putoczki T, Belz GT. Mielke LA, et al. J Immunol. 2013 Oct 15;191(8):4383-91. doi: 10.4049/jimmunol.1301228. Epub 2013 Sep 13. J Immunol. 2013. PMID: 24038093 - Distinct requirements for T-bet in gut innate lymphoid cells.
Sciumé G, Hirahara K, Takahashi H, Laurence A, Villarino AV, Singleton KL, Spencer SP, Wilhelm C, Poholek AC, Vahedi G, Kanno Y, Belkaid Y, O'Shea JJ. Sciumé G, et al. J Exp Med. 2012 Dec 17;209(13):2331-8. doi: 10.1084/jem.20122097. Epub 2012 Dec 3. J Exp Med. 2012. PMID: 23209316 Free PMC article. - Group 3 innate lymphoid cells continuously require the transcription factor GATA-3 after commitment.
Zhong C, Cui K, Wilhelm C, Hu G, Mao K, Belkaid Y, Zhao K, Zhu J. Zhong C, et al. Nat Immunol. 2016 Feb;17(2):169-78. doi: 10.1038/ni.3318. Epub 2015 Nov 23. Nat Immunol. 2016. PMID: 26595886 Free PMC article. - Deciphering the transcriptional switches of innate lymphoid cell programming: the right factors at the right time.
Lim AW, McKenzie AN. Lim AW, et al. Genes Immun. 2015 Apr-May;16(3):177-86. doi: 10.1038/gene.2014.83. Epub 2015 Jan 22. Genes Immun. 2015. PMID: 25611557 Free PMC article. Review. - Diversity and function of group 1 innate lymphoid cells.
Cortez VS, Colonna M. Cortez VS, et al. Immunol Lett. 2016 Nov;179:19-24. doi: 10.1016/j.imlet.2016.07.005. Epub 2016 Jul 6. Immunol Lett. 2016. PMID: 27394699 Free PMC article. Review.
Cited by
- Keeping time in group 3 innate lymphoid cells.
Wang Q, Colonna M. Wang Q, et al. Nat Rev Immunol. 2020 Dec;20(12):720-726. doi: 10.1038/s41577-020-0397-z. Epub 2020 Aug 5. Nat Rev Immunol. 2020. PMID: 32759971 Review. - NK Cells and Other Innate Lymphoid Cells in Hematopoietic Stem Cell Transplantation.
Vacca P, Montaldo E, Croxatto D, Moretta F, Bertaina A, Vitale C, Locatelli F, Mingari MC, Moretta L. Vacca P, et al. Front Immunol. 2016 May 18;7:188. doi: 10.3389/fimmu.2016.00188. eCollection 2016. Front Immunol. 2016. PMID: 27242795 Free PMC article. Review. - ILC2s masquerade as ILC1s to drive chronic disease.
Belz GT. Belz GT. Nat Immunol. 2016 May 19;17(6):611-2. doi: 10.1038/ni.3467. Nat Immunol. 2016. PMID: 27196511 No abstract available. - Immune modules shared by innate lymphoid cells and T cells.
Robinette ML, Colonna M. Robinette ML, et al. J Allergy Clin Immunol. 2016 Nov;138(5):1243-1251. doi: 10.1016/j.jaci.2016.09.006. J Allergy Clin Immunol. 2016. PMID: 27817796 Free PMC article. Review. - Brg1 restrains the pro-inflammatory properties of ILC3s and modulates intestinal immunity.
Qi X, Qiu J, Chang J, Ji Y, Yang Q, Cui G, Sun L, Chai Q, Qin J, Qiu J. Qi X, et al. Mucosal Immunol. 2021 Jan;14(1):38-52. doi: 10.1038/s41385-020-0317-3. Epub 2020 Jul 1. Mucosal Immunol. 2021. PMID: 32612160 Free PMC article.
References
- Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. - PubMed
- Sun Z, et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. - PubMed
- Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, Gruss P. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;25:702–706. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials