Regulation of A20 and other OTU deubiquitinases by reversible oxidation - PubMed (original) (raw)
Regulation of A20 and other OTU deubiquitinases by reversible oxidation
Yogesh Kulathu et al. Nat Commun. 2013.
Abstract
Protein ubiquitination is a highly versatile post-translational modification that regulates as diverse processes as protein degradation and kinase activation. Deubiquitinases hydrolyse ubiquitin modifications from proteins and are hence key regulators of the ubiquitin system. Ovarian tumour deubiquitinases comprise a family of fourteen human enzymes, many of which regulate cellular signalling pathways. Ovarian tumour deubiquitinases are cysteine proteases that cleave polyubiquitin chains in vitro and in cells, but little is currently known about their regulation. Here we show that ovarian tumour deubiquitinases are susceptible to reversible oxidation of the catalytic cysteine residue. High-resolution crystal structures of the catalytic domain of A20 in four different oxidation states reveal that the reversible form of A20 oxidation is a cysteine sulphenic acid intermediate, which is stabilised by the architecture of the catalytic centre. Using chemical tools to detect sulphenic acid intermediates, we show that many ovarian tumour deubiquitinases undergo reversible oxidation upon treatment with H2O2, revealing a new mechanism to regulate deubiquitinase activity.
Figures
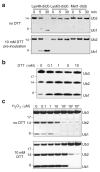
Figure 1. Reversible A20 oxidation in vitro
a) DUB assay of A20 OTU against Lys48- Lys63- and Met1-linked diUb. The DUB assay was carried out either in the absence of reducing agents in the reaction buffer (upper panel) or the DUB was first pre-incubated with 10 mM DTT for 15 min prior to the DUB assay. b) DUB assay of A20 OTU against Lys48-linked triUb was performed with pre-incubation of the OTU in increasing concentrations of DTT. The reaction was carried out in buffer lacking DTT. c) A20 was first incubated with indicated concentrations of H2O2 for 15 min at 23 °C, and 100 U of catalase was added to quench the H2O2. To one half of the sample, DTT was added to a final concentration of 10 mM and incubated at room temperature for 15 min. The activity of the treated A20 was tested in a DUB assay using Lys48-linked triUb as substrate.
Figure 2. Detection of sulphenic acid in reversibly oxidised A20
a) Schematic of the reaction. Structure of A20 (pdb-id 2vfj, ) showing the six ordered Cys residues (green). Treatment of A20 with dimedone derivatives, such as DAz-2, tests for Cys sulphenic acid intermediates, which can be detected by western blotting (as in b) or by mass-spectrometry (as in c) b) Purified A20 was treated with 50 μM H2O2 for 60 min at room temperature and incubated with DMSO or DAz-2. Subsequent biotinylation allows detection of A20 modification by Strepavidin-coupled horseradish peroxidase (Strep-HRP). c) Left, Mass-spectra of intact A20 OTU domain (calculated mass 43452 Da, labelled with black asterisks) without H2O2 treatment. Middle, untreated A20 OTU domain labelled with DAz-2. Right, A20 OTU domain treated with 50 μM H2O2 for 1 h at room temperature, and labelled with DAz-2. Labelling with DAz-2 (195 Da) results in a second peak (labelled with red asterisks) in untreated and H2O2 treated samples (observed mass 43654/43646, respectively). The observed mass corresponds to A20 labelled with 1 molecule of DAz-2. d) Tryptic analysis of unlabelled (left) and labelled (right) A20 reveals the Cys103-containing peptide to be modified with DAz-2 (see Methods). Shown is the sequence of the Cys103 containing tryptic peptide including b- and y-ions (top), extracted ion chromatograms (XIC, middle), and MS/MS spectra after collision induced dissociation (CID, bottom). The MS/MS spectra after CID of precursor ion 1074.36 [M+3H]+3 yielded specific b and y ions pertaining to the peptide with DAz-2 modification at Cys103.
Figure 3. A high-resolution crystal form for A20 OTU
a) Structure of A20 OTU domain (aa 1-366) in _P_1 space group at 1.87 Å resolution. N- and C-terminus and the presumed S1 Ub binding site (from analogy with other OTU complexes, e.g. ) are indicated. Residues of the catalytic triad (Cys103, His256, Asp70) are shown. Dotted lines indicate connectivity of disordered surface loops. b) Structure of the A20 OTU dimer in the asymmetric unit. c) Superposition of A20 OTU from this study (red) and from a previous study (pdb-id 3dkb, ). RMSD between structures is 0.8 Å over 322/352 residues.
Figure 4. The A20 catalytic triad in different oxidation states
a-d) Stereo images of the catalytic Cys and His residues and the Cys-loop in A20. A 2∣Fo∣-∣Fc∣ map (blue) contoured at 1.5 σ, and a ∣Fo∣-∣Fc∣ map (green, thick lines) contoured at 3.5 σ covers shown residues. Left: Molecule A of the asymmetric unit. RIght: Molecule B of the asymmetric unit. a) Structure of A20 in reduced, active state, grown in presence of DTT. b) Structure of A20 with reversible oxidised Cys103 sulphenic acid, from untreated crystals. c) Structure of A20 with irreversibly oxidised Cys103 after incubation with 50 μM H2O2 for 60 min. Cys residues were modelled as sulphinic and sulphonic acid in the two molecules, respectively. d) Structure of A20 after 270 min incubation with 50 μM H2O2, modelled as in c. e) The remaining ordered Cys residues in A20 (Cys54, Cys57, Cys86, Cys200, Cys243) are unaffected even after soaking for 270 min in 50 μM H2O2. Individual Cys residues are shown with 2∣Fo∣-∣Fc∣ map (blue, contoured at 1.5 σ) and ∣Fo∣-∣Fc∣ map (green, contoured at 3.5 σ). There is no un-modelled density at these sites. f) Final, refined models for reduced (A20 SH), reversibly oxidised (A20 SOH) and irreversibly (A20 SO2H, A20 SO3H) oxidised forms of A20. Yellow dotted lines indicate hydrogen bonds formed by oxidised Cys103.
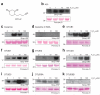
Figure 5. Detection of sulphenylation in different OTU DUBs
a) Chemical structure of DYn-2, which features an alkyne group. b-k) Catalytic domains of different human OTU family members were treated with increasing concentrations of H2O2 for 15 minutes at room temperature. H2O2 was quenched by adding 200 U catalase and the OTU was labelled with 1 μM DYn-2, which was subsequently biotinylated. The labelled proteins were detected by streptavidin-horseradish peroxidase (Strep-HRP) western blot. Ponceau S staining of the membrane confirmed equal loading of proteins. b) A20 OTU domain (aa 1-366), c) Cezanne/OTUD7b (aa 124-438), d) Cezanne/OTUD7b C194A (aa 124-438), e) OTUB1 (aa 1-271, full length), f) OTUD1 (aa 287-481), g) OTUD2 (aa 1-348, full length), h) OTUD3 (aa 52-275), i) OTUD5/DUBA (aa 171-358), j) OTUD6A (aa 128-288), k) OTUD6B (aa 133-293).
Similar articles
- Structure of the A20 OTU domain and mechanistic insights into deubiquitination.
Komander D, Barford D. Komander D, et al. Biochem J. 2008 Jan 1;409(1):77-85. doi: 10.1042/BJ20071399. Biochem J. 2008. PMID: 17961127 - Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20.
Lin SC, Chung JY, Lamothe B, Rajashankar K, Lu M, Lo YC, Lam AY, Darnay BG, Wu H. Lin SC, et al. J Mol Biol. 2008 Feb 15;376(2):526-40. doi: 10.1016/j.jmb.2007.11.092. Epub 2007 Dec 4. J Mol Biol. 2008. PMID: 18164316 Free PMC article. - The biology of A20-like molecules.
Enesa K, Evans P. Enesa K, et al. Adv Exp Med Biol. 2014;809:33-48. doi: 10.1007/978-1-4939-0398-6_3. Adv Exp Med Biol. 2014. PMID: 25302364 Review. - Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity.
Evans PC, Ovaa H, Hamon M, Kilshaw PJ, Hamm S, Bauer S, Ploegh HL, Smith TS. Evans PC, et al. Biochem J. 2004 Mar 15;378(Pt 3):727-34. doi: 10.1042/BJ20031377. Biochem J. 2004. PMID: 14748687 Free PMC article. - Regulation of NF-κB by deubiquitinases.
Harhaj EW, Dixit VM. Harhaj EW, et al. Immunol Rev. 2012 Mar;246(1):107-24. doi: 10.1111/j.1600-065X.2012.01100.x. Immunol Rev. 2012. PMID: 22435550 Free PMC article. Review.
Cited by
- A20/Tumor Necrosis Factor α-Induced Protein 3 in Immune Cells Controls Development of Autoinflammation and Autoimmunity: Lessons from Mouse Models.
Das T, Chen Z, Hendriks RW, Kool M. Das T, et al. Front Immunol. 2018 Feb 21;9:104. doi: 10.3389/fimmu.2018.00104. eCollection 2018. Front Immunol. 2018. PMID: 29515565 Free PMC article. Review. - Diverse Redoxome Reactivity Profiles of Carbon Nucleophiles.
Gupta V, Yang J, Liebler DC, Carroll KS. Gupta V, et al. J Am Chem Soc. 2017 Apr 19;139(15):5588-5595. doi: 10.1021/jacs.7b01791. Epub 2017 Apr 10. J Am Chem Soc. 2017. PMID: 28355876 Free PMC article. - Erasing marks: Functions of plant deubiquitylating enzymes in modulating the ubiquitin code.
Vogel K, Isono E. Vogel K, et al. Plant Cell. 2024 Sep 3;36(9):3057-3073. doi: 10.1093/plcell/koae129. Plant Cell. 2024. PMID: 38656977 Free PMC article. Review. - Chemical proteomics reveals new targets of cysteine sulfinic acid reductase.
Akter S, Fu L, Jung Y, Conte ML, Lawson JR, Lowther WT, Sun R, Liu K, Yang J, Carroll KS. Akter S, et al. Nat Chem Biol. 2018 Nov;14(11):995-1004. doi: 10.1038/s41589-018-0116-2. Epub 2018 Sep 3. Nat Chem Biol. 2018. PMID: 30177848 Free PMC article. - A20 at the Crossroads of Cell Death, Inflammation, and Autoimmunity.
Martens A, van Loo G. Martens A, et al. Cold Spring Harb Perspect Biol. 2020 Jan 2;12(1):a036418. doi: 10.1101/cshperspect.a036418. Cold Spring Harb Perspect Biol. 2020. PMID: 31427375 Free PMC article. Review.
References
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. - PubMed
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. - PubMed
- Kulathu Y, Komander D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 2012;13:508–523. - PubMed
- Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 309756/ERC_/European Research Council/International
- A7403/CRUK_/Cancer Research UK/United Kingdom
- U105192732/MRC_/Medical Research Council/United Kingdom
- MC_UP_1201/6/MRC_/Medical Research Council/United Kingdom
- U.1051.03.019.00001.01(92732)/MRC_/Medical Research Council/United Kingdom
- MC_U105192732/MRC_/Medical Research Council/United Kingdom
- R01 CA174864/CA/NCI NIH HHS/United States
- A2560/CRUK_/Cancer Research UK/United Kingdom
- R01 GM102187/GM/NIGMS NIH HHS/United States
- 14109/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical