The Pacific Ocean virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology - PubMed (original) (raw)
The Pacific Ocean virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology
Bonnie L Hurwitz et al. PLoS One. 2013.
Abstract
Bacteria and their viruses (phage) are fundamental drivers of many ecosystem processes including global biogeochemistry and horizontal gene transfer. While databases and resources for studying function in uncultured bacterial communities are relatively advanced, many fewer exist for their viral counterparts. The issue is largely technical in that the majority (often 90%) of viral sequences are functionally 'unknown' making viruses a virtually untapped resource of functional and physiological information. Here, we provide a community resource that organizes this unknown sequence space into 27 K high confidence protein clusters using 32 viral metagenomes from four biogeographic regions in the Pacific Ocean that vary by season, depth, and proximity to land, and include some of the first deep pelagic ocean viral metagenomes. These protein clusters more than double currently available viral protein clusters, including those from environmental datasets. Further, a protein cluster guided analysis of functional diversity revealed that richness decreased (i) from deep to surface waters, (ii) from winter to summer, (iii) and with distance from shore in surface waters only. These data provide a framework from which to draw on for future metadata-enabled functional inquiries of the vast viral unknown.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
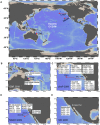
Figure 1. Sampling site map for the POV dataset.
Thirty-two viral metagenomes represent discretely sampled and processed datasets that vary over time and space in the pelagic Pacific Ocean. (A) Overview of sampling sites, (B) GBR – Great Barrier Reef, Australia, near Dunk and Fitzroy Islands. (C) LineP- oceanographic transect off Vancouver Island, British Columbia (D) MBARI- Line67 oceanographic transect off of Monterey Bay, California (E) SIO- Scripps Pier, San Diego, CA. Images were created using Ocean Data View.
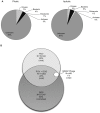
Figure 2. The POV dataset and its place in the viral protein universe.
(A) Summary superkingdom taxonomy statistics for quantitative Pacific Ocean viral metagenomes from 16 photic and 16 aphotic zone samples. Reads were taxonomically assigned based on matches to proteins in SIMAP and curated as described in the methods. (B) Venn diagram representing medium- to large membership PCs documents the relative contributions of the POV, GOS microbial, and SIMAP datasets to the ‘viral protein universe’.
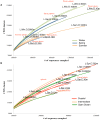
Figure 3. Viral community functional richness based on season and proximity to shore.
Rarefaction analysis of hits to protein clusters from: (A) 11 POV metagenomes from a single LineP open ocean site (station P26) (B) 11 POV metagenomes from LineP stations P4, P12, and P26 from a single research cruise (June 2009). To be conservative, only protein clusters with >20 members were used in these analyses.
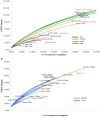
Figure 4. Viral community functional richness in the Pacific Ocean.
Rarefaction analysis of hits to protein clusters from all POV metagenomes in (A) photic zone samples and (B) aphotic zone samples. To be conservative, only protein clusters with >20 members were used in these analyses.
Similar articles
- Modeling ecological drivers in marine viral communities using comparative metagenomics and network analyses.
Hurwitz BL, Westveld AH, Brum JR, Sullivan MB. Hurwitz BL, et al. Proc Natl Acad Sci U S A. 2014 Jul 22;111(29):10714-9. doi: 10.1073/pnas.1319778111. Epub 2014 Jul 7. Proc Natl Acad Sci U S A. 2014. PMID: 25002514 Free PMC article. - Depth-stratified functional and taxonomic niche specialization in the 'core' and 'flexible' Pacific Ocean Virome.
Hurwitz BL, Brum JR, Sullivan MB. Hurwitz BL, et al. ISME J. 2015 Feb;9(2):472-84. doi: 10.1038/ismej.2014.143. Epub 2014 Aug 5. ISME J. 2015. PMID: 25093636 Free PMC article. - Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses.
Roux S, Brum JR, Dutilh BE, Sunagawa S, Duhaime MB, Loy A, Poulos BT, Solonenko N, Lara E, Poulain J, Pesant S, Kandels-Lewis S, Dimier C, Picheral M, Searson S, Cruaud C, Alberti A, Duarte CM, Gasol JM, Vaqué D; Tara Oceans Coordinators; Bork P, Acinas SG, Wincker P, Sullivan MB. Roux S, et al. Nature. 2016 Sep 29;537(7622):689-693. doi: 10.1038/nature19366. Epub 2016 Sep 21. Nature. 2016. PMID: 27654921 - Viruses in Soil Ecosystems: An Unknown Quantity Within an Unexplored Territory.
Williamson KE, Fuhrmann JJ, Wommack KE, Radosevich M. Williamson KE, et al. Annu Rev Virol. 2017 Sep 29;4(1):201-219. doi: 10.1146/annurev-virology-101416-041639. Annu Rev Virol. 2017. PMID: 28961409 Review. - Metagenomics Sheds Light on the Ecology of Marine Microbes and Their Viruses.
Coutinho FH, Gregoracci GB, Walter JM, Thompson CC, Thompson FL. Coutinho FH, et al. Trends Microbiol. 2018 Nov;26(11):955-965. doi: 10.1016/j.tim.2018.05.015. Epub 2018 Jun 21. Trends Microbiol. 2018. PMID: 29937307 Review.
Cited by
- Twelve previously unknown phage genera are ubiquitous in global oceans.
Holmfeldt K, Solonenko N, Shah M, Corrier K, Riemann L, Verberkmoes NC, Sullivan MB. Holmfeldt K, et al. Proc Natl Acad Sci U S A. 2013 Jul 30;110(31):12798-803. doi: 10.1073/pnas.1305956110. Epub 2013 Jul 15. Proc Natl Acad Sci U S A. 2013. PMID: 23858439 Free PMC article. - The Fennoscandian Shield deep terrestrial virosphere suggests slow motion 'boom and burst' cycles.
Holmfeldt K, Nilsson E, Simone D, Lopez-Fernandez M, Wu X, de Bruijn I, Lundin D, Andersson AF, Bertilsson S, Dopson M. Holmfeldt K, et al. Commun Biol. 2021 Mar 8;4(1):307. doi: 10.1038/s42003-021-01810-1. Commun Biol. 2021. PMID: 33686191 Free PMC article. - Targeted diversity generation by intraterrestrial archaea and archaeal viruses.
Paul BG, Bagby SC, Czornyj E, Arambula D, Handa S, Sczyrba A, Ghosh P, Miller JF, Valentine DL. Paul BG, et al. Nat Commun. 2015 Mar 23;6:6585. doi: 10.1038/ncomms7585. Nat Commun. 2015. PMID: 25798780 Free PMC article. - Virus-Host Interactions and Genetic Diversity of Antarctic Sea Ice Bacteriophages.
Demina TA, Luhtanen AM, Roux S, Oksanen HM. Demina TA, et al. mBio. 2022 Jun 28;13(3):e0065122. doi: 10.1128/mbio.00651-22. Epub 2022 May 9. mBio. 2022. PMID: 35532161 Free PMC article. - Newly identified HMO-2011-type phages reveal genomic diversity and biogeographic distributions of this marine viral group.
Qin F, Du S, Zhang Z, Ying H, Wu Y, Zhao G, Yang M, Zhao Y. Qin F, et al. ISME J. 2022 May;16(5):1363-1375. doi: 10.1038/s41396-021-01183-7. Epub 2022 Jan 12. ISME J. 2022. PMID: 35022515 Free PMC article.
References
- Falkowski P, Fenchel T, Delong E (2008) The microbial engines that drive Earth’s biogeochemical cycles. Science: 1034–1039. - PubMed
- Weinbauer MG (2004) Ecology of prokaryotic viruses. FEMS Microbiology Reviews 28: 127–181. - PubMed
- Fuhrman JA (1999) Marine viruses and their biogeochemical and ecological effects. Nature 399: 541–548. - PubMed
- Rohwer F, Thurber RV (2009) Viruses manipulate the marine environment. Nature 459: 207–212. - PubMed
Publication types
MeSH terms
Grants and funding
Sequencing was provided by the Department of Energy Joint Genome Institute Community Sequencing Program and the Gordon and Betty Moore Foundation Marine Microbial Initiative, while funding was from National Science Foundation (DBI-0850105 and OCE-0961947), Biosphere 2, BIO5 and Gordon and Betty Moore Foundation grants. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources