Discovery of functional toxin/antitoxin systems in bacteria by shotgun cloning - PubMed (original) (raw)
Discovery of functional toxin/antitoxin systems in bacteria by shotgun cloning
Hila Sberro et al. Mol Cell. 2013.
Abstract
Toxin-antitoxin (TA) modules, composed of a toxic protein and a counteracting antitoxin, play important roles in bacterial physiology. We examined the experimental insertion of 1.5 million genes from 388 microbial genomes into an Escherichia coli host using more than 8.5 million random clones. This revealed hundreds of genes (toxins) that could only be cloned when the neighboring gene (antitoxin) was present on the same clone. Clustering of these genes revealed TA families widespread in bacterial genomes, some of which deviate from the classical characteristics previously described for such modules. Introduction of these genes into E. coli validated that the toxin toxicity is mitigated by the antitoxin. Infection experiments with T7 phage showed that two of the new modules can provide resistance against phage. Moreover, our experiments revealed an "antidefense" protein in phage T7 that neutralizes phage resistance. Our results expose active fronts in the arms race between bacteria and phage.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
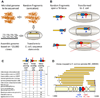
Figure 1. Data derived from whole-genome shotgun sequencing exposes toxin-antitoxin pairs
(A) The "Sanger" based process of DNA sequencing involves random genome fragmentation and transformation of DNA fragments into E. coli. (B) In a DNA locus spanning a toxin/antitoxin (TA) gene pair, random fragmentation leaving the toxin detached from its cognate antitoxin leads to E. coli growth arrest, whereas a fragment containing both genes, or the antitoxin alone, will be propagated and sequenced. (C) A known family of toxin/antitoxin gene pairs, of the VapBC type, was found in 11 of the analyzed genomes. In 7/11 cases the gene pair follows the "TA cloning pattern", significantly higher than the number expected by chance (p=4×10−3). (D) The vapBC locus in Haemophilus somnus 129T (accession NC_008309, locus tags HS_1769-HS_1770). Shown are clones (brown) mapped to the reference genome at that locus. Clones covering the antitoxin but not the toxin are numbered. Only five of the 22 clones covering both toxin and antitoxin are shown.
Figure 2. Workflow for systematic discovery of families of toxin/antitoxin associated with anti-phage defense
Families are divided to "known", representing families that were previously experimentally established as TA systems; "not previously validated", representing families for which there was so far no experimental support of being TA systems; and "complex genomic environment", representing families of genes pairs embedded in a larger operon. Cumulative number of gene pairs belonging to each subset of families is indicated.
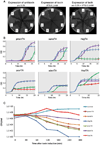
Figure 3. Properties of novel, experimentally verified TA systems
(A) growth of bacteria when only antitoxin is induced (left), only toxin is induced (middle) and both are induced together (right). Toxin and antitoxin were cloned on pRSF (IPTG inducible promoter) and pBAD (arabinose inducible), respectively, in E. coli BL21(DE3)pLysS. C, control bacteria with empty plasmids; 1-pmen system; 2-sana system; 3-psyr system; 4-rleg system; 5-sden system; 6-hhal system. (B) Kinetics of E. coli BL21(DE3)pLysS growth when toxin and antitoxin are co-expressed simultaneously (purple), alone (red and blue for toxin and antitoxin, respectively) or when antitoxin is induced 2.5 hr following toxin induction (green). Kinetic measurements were performed on biological triplicates in technical duplicates. Error bars represent standard deviation. (C) Viability assays for cells following exposure to toxin. Transcription of toxin was induced by 100µM IPTG. At increasing time points following toxin induction (30, 60, 120, 180, 240 and 300 mins) cells were plated on LB-plates containing 0.3% arabinose and no IPTG, to activate antitoxin expression. Colony forming units (CFUs) were determined by colony counting.
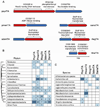
Figure 4. Properties of families experimentally validated in this study
(A) Operon and domain organization of the validated families. Representative pair of each family is shown. For each pair, red and blue genes denote toxin and antitoxin, respectively. (B) Distribution of TA families among different bacterial phyla and (C) human associated bacteria. Number of instances of each system within a phylum/bacterial species is indicated, with darker colors indicating higher number of instances (the phylogenetic distribution of the hhalTA family does not appear here as it was described previously by Makarova et al (2009)).
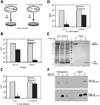
Figure 5. T7 Gp4.5 antagonizes abortive infection and interacts with Lon protease
(A) Illustration of the plaque forming unit (PFU) assays on E. coli harboring toxin/antitoxin systems. Efficiency of plating (EOP) was calculated by dividing the number of PFUs obtained for a bacterial lawn expressing the toxin + antitoxin by the number of PFUs obtained on a lawn expressing the antitoxin alone. Bars in panels B, C and E represent average ± SD of three independent EOP experiments. (B) EOP experiments with E. coli harboring the sanaTA system when infected by WT T7 (left) or by T7Δ4.5 (right). Error bars represent standard deviation between replicates. (C) EOP experiments with WT E. coli (left) and E. coli expressing Gp4.5 (right), when infected by T7Δ4.3Δ4.5Δ4.7. Error bars represent standard deviation between replicates. (D) EOP experiments with WT E. coli (left) and E. coli lacking lon (right), when infected by T7Δ4.3Δ4.5Δ4.7. Error bars represent standard deviation between replicates. (E) Co-immunoprecipitation (co-IP) of Lon and Gp4.5. The E. coli Lon protease was Flag-tagged at the N-terminus and expressed within E. coli BL21(DE3) with or without co-expression of gene 4.5. Samples were analyzed by 15% SDS-PAGE. Three left lanes, total soluble proteins; three lanes on the right, following immunoprecipitation with anti-Flag antibody. Co-IP of Lon and Gp4.5 was validated by mass spectrometry analysis. Numbers on the left denote protein marker sizes in kDa. (F) Western blot analysis on reciprocal co-IP with flag-tagged Gp4.5 and his-tagged Lon. Immunoprecipitation was done with anti-Flag antibody.
Similar articles
- Activation of Toxin-Antitoxin System Toxins Suppresses Lethality Caused by the Loss of σE in Escherichia coli.
Daimon Y, Narita S, Akiyama Y. Daimon Y, et al. J Bacteriol. 2015 Jul;197(14):2316-24. doi: 10.1128/JB.00079-15. Epub 2015 Apr 27. J Bacteriol. 2015. PMID: 25917909 Free PMC article. - Global Analysis of the Specificities and Targets of Endoribonucleases from Escherichia coli Toxin-Antitoxin Systems.
Culviner PH, Nocedal I, Fortune SM, Laub MT. Culviner PH, et al. mBio. 2021 Oct 26;12(5):e0201221. doi: 10.1128/mBio.02012-21. Epub 2021 Sep 21. mBio. 2021. PMID: 34544284 Free PMC article. - A novel family of Escherichia coli toxin-antitoxin gene pairs.
Brown JM, Shaw KJ. Brown JM, et al. J Bacteriol. 2003 Nov;185(22):6600-8. doi: 10.1128/JB.185.22.6600-6608.2003. J Bacteriol. 2003. PMID: 14594833 Free PMC article. - Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response.
Wang X, Wood TK. Wang X, et al. Appl Environ Microbiol. 2011 Aug 15;77(16):5577-83. doi: 10.1128/AEM.05068-11. Epub 2011 Jun 17. Appl Environ Microbiol. 2011. PMID: 21685157 Free PMC article. Review. - Divergently overlapping cis-encoded antisense RNA regulating toxin-antitoxin systems from E. coli: hok/sok, ldr/rdl, symE/symR.
Kawano M. Kawano M. RNA Biol. 2012 Dec;9(12):1520-7. doi: 10.4161/rna.22757. Epub 2012 Nov 6. RNA Biol. 2012. PMID: 23131729 Review.
Cited by
- Biology and evolution of bacterial toxin-antitoxin systems.
Jurėnas D, Fraikin N, Goormaghtigh F, Van Melderen L. Jurėnas D, et al. Nat Rev Microbiol. 2022 Jun;20(6):335-350. doi: 10.1038/s41579-021-00661-1. Epub 2022 Jan 2. Nat Rev Microbiol. 2022. PMID: 34975154 Review. - Revenge of the phages: defeating bacterial defences.
Samson JE, Magadán AH, Sabri M, Moineau S. Samson JE, et al. Nat Rev Microbiol. 2013 Oct;11(10):675-87. doi: 10.1038/nrmicro3096. Epub 2013 Aug 27. Nat Rev Microbiol. 2013. PMID: 23979432 Review. - Bacteriophage strategies for overcoming host antiviral immunity.
Gao Z, Feng Y. Gao Z, et al. Front Microbiol. 2023 Jun 8;14:1211793. doi: 10.3389/fmicb.2023.1211793. eCollection 2023. Front Microbiol. 2023. PMID: 37362940 Free PMC article. Review. - The Extraintestinal Pathogenic Escherichia coli Factor RqlI Constrains the Genotoxic Effects of the RecQ-Like Helicase RqlH.
Russell CW, Mulvey MA. Russell CW, et al. PLoS Pathog. 2015 Dec 4;11(12):e1005317. doi: 10.1371/journal.ppat.1005317. eCollection 2015 Dec. PLoS Pathog. 2015. PMID: 26636713 Free PMC article. - Evolutionary Genomics of Defense Systems in Archaea and Bacteria.
Koonin EV, Makarova KS, Wolf YI. Koonin EV, et al. Annu Rev Microbiol. 2017 Sep 8;71:233-261. doi: 10.1146/annurev-micro-090816-093830. Epub 2017 Jun 28. Annu Rev Microbiol. 2017. PMID: 28657885 Free PMC article. Review.
References
- Bergh O, Borsheim KY, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. - PubMed
- Bernard P, Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol. 1992;226:735–745. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources