Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for β-barrel assembly in Escherichia coli - PubMed (original) (raw)
Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for β-barrel assembly in Escherichia coli
Nathan W Rigel et al. Proc Natl Acad Sci U S A. 2013.
Abstract
In gram-negative bacteria, integral outer membrane β-barrel proteins (OMPs) are assembled by the beta-barrel assembly machine (Bam) complex. The essential components of this complex are the OMP BamA [which contains a carboxyl-terminal β-barrel and an amino-terminal periplasmic module composed of five polypeptide transport associated (POTRA) domains] and the lipoprotein BamD. In Escherichia coli, the Bam complex also contains three nonessential lipoproteins (BamBCE), all of which require the barrel-proximal POTRA domain (P5) for stable interactions with BamA. We have previously reported that the BamA β-barrel assumes two different conformations. A method for conformation-specific labeling of BamA described here reveals that these conformers reflect the degree of surface exposure of the conserved sixth extracellular loop (L6). L6 is surface accessible in one conformation but not in the other, likely because it occupies the lumen of the BamA β-barrel in the latter case. A gain-of-function mutation that promotes Bam activity (bamDR197L) and a loss-of-function mutation that decreases the activity of Bam (ΔbamE) both favor surface exposure of BamA L6, suggesting that BamD and BamE normally act to control L6 exposure through opposing functions. These results, along with the synthetic lethality of the bamDR197L ΔbamE double mutant, imply a cyclic mechanism in which the Bam lipoproteins regulate the conformation of BamA during the OMP assembly reaction. Our results further suggest that BamDE controls L6 exposure via conformational signals transmitted through P5 to L6.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
BamA exists in two conformations. (A) Cell pellets from wild-type, bamDR197L, and Δ_bamE_ strains were treated with proteinase K where indicated. The protease was inactivated by treatment with PMSF, and samples were analyzed by SDS/PAGE and immunoblot using anti-BamA and anti-MBP antibodies. MBP serves as a loading control to ensure that protease does not enter the periplasm. (B) Wild-type, bamDR197L, and Δ_bamE_ cells were processed by gentle lysis and treated at the indicated temperatures. Samples were analyzed by SDS/PAGE and immunoblot using anti-BamA and anti-BamC antibodies.
Fig. 2.
BamA L6 is surface-exposed and contains an intramolecular disulfide bond. (A) Cells from wild-type or bamA cysteine mutants were labeled with Mal-PEG as described in
SI Materials and Methods
either with or without DTT pretreatment. (B) Folding of BamA was assessed in wild-type (bamA CC) and double mutant (bamA SS) cells by performing SDS/PAGE under reducing (Lower) or nonreducing (Upper) conditions. β-Mercaptoethanol (β-ME) was used where indicated as the reducing agent. (C) Labeling as described in A was performed on wild-type, bamDR197L, or Δ_bamE_ strains. Blots were probed with anti-BamA antibodies.
Fig. 3.
Genetic interactions between bamDR197L and Δ_bamE_. (A) Growth of wild-type (square), bamDR197L (circle), Δ_bamE::cam_ (diamond), and bamDR197L Δ_bamE::cam_ (triangle) strains at 37 °C was monitored by measuring OD600. All strains were grown to stationary phase at 30 °C and then diluted 1:1,000 in fresh LB and grown at 37 °C. (B) OMP levels were determined by immunoblot in cell extracts of wild-type, bamDR197L, Δ_bamE::cam_, and bamDR197L Δ_bamE::cam_ strains grown at 30 °C.
Fig. 4.
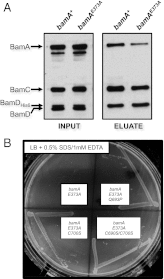
BamA L6 and BamDE communicate via POTRA5. (A) A His-tagged variant of BamD was expressed in bamA + or bamA E373A strains to enable copurification of the Bam complex. Extracts of each strain were purified using a Ni-NTA affinity resin and eluted with imidazole. Input and eluted fractions were analyzed by SDS/PAGE and immunoblot with antibodies that recognize BamA, BamC, or BamD. (B) Strains carrying bamAE373A, bamAE373A+Q693P, bamAE373A+C700S, or bamAE373A+C690S+C700S mutations were streaked onto LB agar plates supplemented with 0.5% SDS and 1.0 mM EDTA. The image was captured after overnight growth at 37 °C.
Fig. 5.
Mutations in BamA L6 can suppress the synthetically lethal bamAE373A bamB8 double mutant. Immunoblot analysis of extracts from the indicated strains were prepared as described in Materials and Methods. Extracts were probed with anti-BamA, anti-BamC, and anti-LamB antibodies.
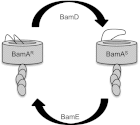
Fig. 6.
Conformational dynamics of the Bam complex. BamA cycles between two different conformations that involve movement of the conserved L6. Interconversion between the protease-resistant, Mal-PEG inaccessible form (BamAR) and the protease-sensitive, Mal-PEG accessible form (BamAS) is regulated by BamDE. In strains carrying gain-of-function bamDR197L and loss-of-function Δ_bamE_ mutations, the equilibrium is shifted to the right.
Similar articles
- Functions of the BamBCDE Lipoproteins Revealed by Bypass Mutations in BamA.
Hart EM, Silhavy TJ. Hart EM, et al. J Bacteriol. 2020 Oct 8;202(21):e00401-20. doi: 10.1128/JB.00401-20. Print 2020 Oct 8. J Bacteriol. 2020. PMID: 32817097 Free PMC article. - The Synthetic Phenotype of Δ_bamB_ Δ_bamE_ Double Mutants Results from a Lethal Jamming of the Bam Complex by the Lipoprotein RcsF.
Hart EM, Gupta M, Wühr M, Silhavy TJ. Hart EM, et al. mBio. 2019 May 21;10(3):e00662-19. doi: 10.1128/mBio.00662-19. mBio. 2019. PMID: 31113901 Free PMC article. - Improper Coordination of BamA and BamD Results in Bam Complex Jamming by a Lipoprotein Substrate.
Tata M, Konovalova A. Tata M, et al. mBio. 2019 May 21;10(3):e00660-19. doi: 10.1128/mBio.00660-19. mBio. 2019. PMID: 31113900 Free PMC article. - The β-Barrel Assembly Machinery Complex.
Leyton DL, Belousoff MJ, Lithgow T. Leyton DL, et al. Methods Mol Biol. 2015;1329:1-16. doi: 10.1007/978-1-4939-2871-2_1. Methods Mol Biol. 2015. PMID: 26427672 Review. - Augmenting β-augmentation: structural basis of how BamB binds BamA and may support folding of outer membrane proteins.
Heuck A, Schleiffer A, Clausen T. Heuck A, et al. J Mol Biol. 2011 Mar 11;406(5):659-66. doi: 10.1016/j.jmb.2011.01.002. Epub 2011 Jan 12. J Mol Biol. 2011. PMID: 21236263 Review.
Cited by
- The translocation assembly module (TAM) catalyzes the assembly of bacterial outer membrane proteins in vitro.
Wang X, Nyenhuis SB, Bernstein HD. Wang X, et al. bioRxiv [Preprint]. 2024 Jun 20:2024.06.20.599893. doi: 10.1101/2024.06.20.599893. bioRxiv. 2024. PMID: 39372782 Free PMC article. Updated. Preprint. - The translocation assembly module (TAM) catalyzes the assembly of bacterial outer membrane proteins in vitro.
Wang X, Nyenhuis SB, Bernstein HD. Wang X, et al. Nat Commun. 2024 Aug 23;15(1):7246. doi: 10.1038/s41467-024-51628-8. Nat Commun. 2024. PMID: 39174534 Free PMC article. - Targeting BamA, the essential component of the Acinetobacter baumannii β-barrel assembly machinery, hinders its ability to adhere to and invade human alveolar basal epithelial cell line.
Momeni M, Fekrirad Z, Jalali Nadoushan M, Rasooli I. Momeni M, et al. Heliyon. 2024 Jul 10;10(14):e34371. doi: 10.1016/j.heliyon.2024.e34371. eCollection 2024 Jul 30. Heliyon. 2024. PMID: 39108912 Free PMC article. - In Vivo Disulfide-Bond Crosslinking to Study β-Barrel Membrane Protein Interactions, Dynamicity, and Folding Intermediates.
Doyle MT. Doyle MT. Methods Mol Biol. 2024;2778:101-115. doi: 10.1007/978-1-0716-3734-0_7. Methods Mol Biol. 2024. PMID: 38478274 - ATP-independent assembly machinery of bacterial outer membranes: BAM complex structure and function set the stage for next-generation therapeutics.
George A, Patil AG, Mahalakshmi R. George A, et al. Protein Sci. 2024 Feb;33(2):e4896. doi: 10.1002/pro.4896. Protein Sci. 2024. PMID: 38284489 Review.
References
- Wu T, et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121(2):235–245. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM093768/GM/NIGMS NIH HHS/United States
- R01 GM034821/GM/NIGMS NIH HHS/United States
- R37 GM034821/GM/NIGMS NIH HHS/United States
- F32 GM093768/GM/NIGMS NIH HHS/United States
- GM34821/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous