Single-cell and population level viral infection dynamics revealed by phageFISH, a method to visualize intracellular and free viruses - PubMed (original) (raw)
. 2013 Aug;15(8):2306-18.
doi: 10.1111/1462-2920.12100. Epub 2013 Mar 14.
Affiliations
- PMID: 23489642
- PMCID: PMC3884771
- DOI: 10.1111/1462-2920.12100
Free PMC article
Single-cell and population level viral infection dynamics revealed by phageFISH, a method to visualize intracellular and free viruses
Elke Allers et al. Environ Microbiol. 2013 Aug.
Free PMC article
Abstract
Microbes drive the biogeochemical cycles that fuel planet Earth, and their viruses (phages) alter microbial population structure, genome repertoire, and metabolic capacity. However, our ability to understand and quantify phage-host interactions is technique-limited. Here, we introduce phageFISH - a markedly improved geneFISH protocol that increases gene detection efficiency from 40% to > 92% and is optimized for detection and visualization of intra- and extracellular phage DNA. The application of phageFISH to characterize infection dynamics in a marine podovirus-gammaproteobacterial host model system corroborated classical metrics (qPCR, plaque assay, FVIC, DAPI) and outperformed most of them to reveal new biology. PhageFISH detected both replicating and encapsidated (intracellular and extracellular) phage DNA, while simultaneously identifying and quantifying host cells during all stages of infection. Additionally, phageFISH allowed per-cell relative measurements of phage DNA, enabling single-cell documentation of infection status (e.g. early vs late stage infections). Further, it discriminated between two waves of infection, which no other measurement could due to population-averaged signals. Together, these findings richly characterize the infection dynamics of a novel model phage-host system, and debut phageFISH as a much-needed tool for studying phage-host interactions in the laboratory, with great promise for environmental surveys and lineage-specific population ecology of free phages.
© 2013 John Wiley & Sons Ltd and Society for Applied Microbiology.
Figures
Figure 1
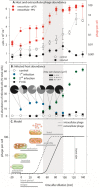
One-step growth infection dynamics of a marine virus–host system.A. Host abundance as total cell counts and extracellular virus abundance as virus abundance per cell by gene presence (qPCR) and plaque assays (PFU).B. Relative abundance of phage containing cells as FVIC by TEM count (black squares) and phageFISH count (coloured circles); pie charts indicate area size distribution among phage signals as determined by phageFISH.C. One-step growth model for phage PSA-HP1 and its host H100.Error bars indicate standard deviation except for FVIC data in which error bars indicate 95% confidence intervals. All data are based on measurements from two biological replicates. All data at T-19 are corrected by a 1/100 factor to be comparable to values measured after dilution of cultures.
Figure 2
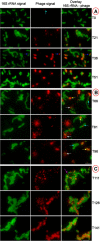
Progression of infection over time: epifluorescence micrographs of virus-infected and uninfected host cells after phageFISH.A: T0–T51, B: T66–T96, C: T111–T46.Left: host only. Centre: virus only. Right column: overlay of host cells in green (Alexa488) and virus in red (Alexa594).Colour designation for arrows: white = cell lysis and release of free phages, cyan = new infection, blue = rRNA and phage localization, yellow = advanced infection, violet = infection by two phage. The scale bar indicates 5 μm.
Figure 3
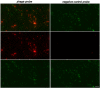
Epifluorescence micrograph of free virus after phageFISH (red, Alexa594) and SYBR staining (green).On the left: probe targeting a viral gene, on the right: negative control gene probe.Top: overlay of SYBR Green and phageFISH, centre: phageFISH only, bottom: SYBR Green only. Scale bar indicates 2 μm.
Similar articles
- Variably lytic infection dynamics of large Bacteroidetes podovirus phi38:1 against two Cellulophaga baltica host strains.
Dang VT, Howard-Varona C, Schwenck S, Sullivan MB. Dang VT, et al. Environ Microbiol. 2015 Nov;17(11):4659-71. doi: 10.1111/1462-2920.13009. Epub 2015 Sep 16. Environ Microbiol. 2015. PMID: 26248067 - Isolation and Genome Sequencing of a Novel Pseudoalteromonas Phage PH1.
Liu Z, Wang M, Meng X, Li Y, Wang D, Jiang Y, Shao H, Zhang Y. Liu Z, et al. Curr Microbiol. 2017 Feb;74(2):212-218. doi: 10.1007/s00284-016-1175-9. Epub 2016 Dec 9. Curr Microbiol. 2017. PMID: 27942842 - Contrasting life strategies of viruses that infect photo- and heterotrophic bacteria, as revealed by viral tagging.
Deng L, Gregory A, Yilmaz S, Poulos BT, Hugenholtz P, Sullivan MB. Deng L, et al. mBio. 2012 Oct 30;3(6):e00373-12. doi: 10.1128/mBio.00373-12. mBio. 2012. PMID: 23111870 Free PMC article. - Emerging methods to study bacteriophage infection at the single-cell level.
Dang VT, Sullivan MB. Dang VT, et al. Front Microbiol. 2014 Dec 23;5:724. doi: 10.3389/fmicb.2014.00724. eCollection 2014. Front Microbiol. 2014. PMID: 25566233 Free PMC article. Review. - Phage puppet masters of the marine microbial realm.
Breitbart M, Bonnain C, Malki K, Sawaya NA. Breitbart M, et al. Nat Microbiol. 2018 Jul;3(7):754-766. doi: 10.1038/s41564-018-0166-y. Epub 2018 Jun 4. Nat Microbiol. 2018. PMID: 29867096 Review.
Cited by
- Droplet Digital PCR for Estimating Absolute Abundances of Widespread Pelagibacter Viruses.
Martinez-Hernandez F, Garcia-Heredia I, Lluesma Gomez M, Maestre-Carballa L, Martínez Martínez J, Martinez-Garcia M. Martinez-Hernandez F, et al. Front Microbiol. 2019 Jun 12;10:1226. doi: 10.3389/fmicb.2019.01226. eCollection 2019. Front Microbiol. 2019. PMID: 31244789 Free PMC article. - Acidianus Tailed Spindle Virus: a New Archaeal Large Tailed Spindle Virus Discovered by Culture-Independent Methods.
Hochstein RA, Amenabar MJ, Munson-McGee JH, Boyd ES, Young MJ. Hochstein RA, et al. J Virol. 2016 Jan 13;90(7):3458-68. doi: 10.1128/JVI.03098-15. J Virol. 2016. PMID: 26763997 Free PMC article. - Quantification of diverse virus populations in the environment using the polony method.
Baran N, Goldin S, Maidanik I, Lindell D. Baran N, et al. Nat Microbiol. 2018 Jan;3(1):62-72. doi: 10.1038/s41564-017-0045-y. Epub 2017 Oct 30. Nat Microbiol. 2018. PMID: 29085077 Free PMC article. - FISH Variants.
Guimarães NM, Azevedo NF, Almeida C. Guimarães NM, et al. Methods Mol Biol. 2021;2246:17-33. doi: 10.1007/978-1-0716-1115-9_2. Methods Mol Biol. 2021. PMID: 33576980 Review. - Classification and quantification of bacteriophage taxa in human gut metagenomes.
Waller AS, Yamada T, Kristensen DM, Kultima JR, Sunagawa S, Koonin EV, Bork P. Waller AS, et al. ISME J. 2014 Jul;8(7):1391-402. doi: 10.1038/ismej.2014.30. Epub 2014 Mar 13. ISME J. 2014. PMID: 24621522 Free PMC article.
References
- Adams MK. Bacteriophages. New York, USA: Interscience Publ; 1959.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources