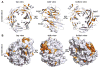Structural and functional studies of the 252 kDa nucleoporin ELYS reveal distinct roles for its three tethered domains - PubMed (original) (raw)
Structural and functional studies of the 252 kDa nucleoporin ELYS reveal distinct roles for its three tethered domains
Silvija Bilokapic et al. Structure. 2013.
Abstract
In metazoa, the nuclear envelope (NE), together with the embedded nuclear pore complexes (NPCs), breaks down and reassembles during cell division. It is suggested that ELYS, a nucleoporin, binds to chromatin in an initial step of postmitotic NPC assembly and subsequently recruits the essential Y-subcomplex, the major scaffolding unit of the NPC. Here, we show that ELYS contains three domains: an N-terminal β-propeller domain, a central α-helical domain, and a C-terminal disordered region. While the disordered region is responsible for the interactions with chromatin, the two preceding domains synergistically mediate tethering to the NPC. We present the crystal structure of the seven-bladed β-propeller domain at 1.9 Å resolution. Analysis of the β-propeller surface reveals the regions that are required for NPC anchorage. We discuss the possible roles of ELYS in the context of the NPC scaffold architecture.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
Figure 1. Domain organization of ELYS
Secondary structure prediction methods suggest three domains in higher eukaryotes, with an N-terminal β-stranded domain (NTD), a central α-helical domain (CHD) and a disordered C-terminal region (CTR). In unicellular eukaryotes, i.e. S. pombe, only the CHD is conserved in ELYS homologs. The constructs used for crystallization are denoted with black bars. The residue numbering is based on UniProt entries Q8CJF7 (M. musculus), Q8WYP5 (H. sapiens), and O94384 (S. pombe).
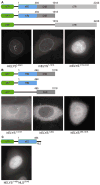
Figure 2. In vivo localization of metazoan ELYS domains
(A) Domain organization of the GFP-labeled mELYS constructs and their localization in HeLa cells. Full-length mELYS (residues 1–2243) shows nuclear rim staining and intra-nuclear puncta. The construct containing both structured domains (NTD and CHD, residues 1–1018) also localize to the nuclear rim, while the disordered region (CTR, residues 1019–2243) shows diffuse nuclear GFP-signal. (B) Domain organization of the GFP-labeled hELYS constructs and their localization in HeLa cells. The constructs containing only the NTD (residues 1–494) or the CHD (residues 495–1018) fail to localize to the nuclear rim in HeLa cells, showing that in metazoa both domains (residues 1–1018) are required for interactions with the NPC. (C) Fusion of the classical SV40 NLS to the C terminus of the hELYS NTD induces nuclear translocation but the domain does not concentrate at the nuclear rim.
Figure 3. Structural analysis of mELYS NTD
(A) Cartoon representation of the mELYS β–propeller domain with the blades labeled from 1 to 7. As a reference, the strands of blade 3 are labeled A–D, with the non-canonical 5th strand labeled D′. The β–propeller blades are colored in blue, loops are in white and the secondary structure insertions within the loops are shown in orange. (B) Comparison of the β–propeller blades reveals long loops that decorate the structural core and contribute about 40% of the total mass of the 494 residue mELYS NTD. See also Figure S1.
Figure 4. Conserved features of the mELYS β–propeller
(A) Cartoon representation of mELYS NTD from three different perspectives, related by rotation around the vertical axis as indicated. Conservation within the mELYS NTD is gradient-colored from gray (not conserved) to dark orange (most conserved) based on the alignment in Figure S3. The majority of conserved residues are structurally important as they build the core scaffold of the β–propeller. (B) Surface representation showing conservation of residues on the β–propeller periphery. Color scheme as in A. The two conserved loops building the NPC binding interface are outlined with a black dotted line. See also Figure S3.
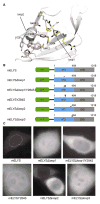
Figure 5. Mutations in the conserved β–propeller loops abolish proper NPC localization of mELYS
(A) Cartoon representation of mELYS NTD with the residues in the conserved loops shown as sticks. Loop1 residues are highlighted in yellow and loop2 residues in pink. Residues forming a hydrophobic pocket beneath loop2 are shown in gray. (B) Domain organization of the GFP-tagged mELYS mutants with the positions of the mutations indicated by red bars (loop deletions or multiple mutations) or red arrowheads (single mutations). (C) Confocal fluorescence microscopy of HeLa cells expressing GFP-tagged mELYS proteins. Wild-type mELYS properly localizes to the nuclear envelope. Deletion of the disordered loop3 (control, residues 458–476), as well as milder mutations within the binding region show nuclear rim staining. The mELYSΔloop1/Y284S and mELYSloop2mut mutants abolish nuclear rim localization. See also Figure S4.
Similar articles
- Structural and biochemical analyses of the nuclear pore complex component ELYS identify residues responsible for nucleosome binding.
Kobayashi W, Takizawa Y, Aihara M, Negishi L, Ishii H, Kurumizaka H. Kobayashi W, et al. Commun Biol. 2019 May 3;2:163. doi: 10.1038/s42003-019-0385-7. eCollection 2019. Commun Biol. 2019. PMID: 31069272 Free PMC article. - The nucleoporin ELYS/Mel28 regulates nuclear envelope subdomain formation in HeLa cells.
Clever M, Funakoshi T, Mimura Y, Takagi M, Imamoto N. Clever M, et al. Nucleus. 2012 Mar 1;3(2):187-99. doi: 10.4161/nucl.19595. Epub 2012 Mar 1. Nucleus. 2012. PMID: 22555603 Free PMC article. - The Role of Nucleoporin Elys in Nuclear Pore Complex Assembly and Regulation of Genome Architecture.
Shevelyov YY. Shevelyov YY. Int J Mol Sci. 2020 Dec 13;21(24):9475. doi: 10.3390/ijms21249475. Int J Mol Sci. 2020. PMID: 33322130 Free PMC article. Review. - Architecture of the cytoplasmic face of the nuclear pore.
Bley CJ, Nie S, Mobbs GW, Petrovic S, Gres AT, Liu X, Mukherjee S, Harvey S, Huber FM, Lin DH, Brown B, Tang AW, Rundlet EJ, Correia AR, Chen S, Regmi SG, Stevens TA, Jette CA, Dasso M, Patke A, Palazzo AF, Kossiakoff AA, Hoelz A. Bley CJ, et al. Science. 2022 Jun 10;376(6598):eabm9129. doi: 10.1126/science.abm9129. Epub 2022 Jun 10. Science. 2022. PMID: 35679405 Free PMC article. - Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: commonalities, differences, challenges.
Leksa NC, Schwartz TU. Leksa NC, et al. Nucleus. 2010 Jul-Aug;1(4):314-8. doi: 10.4161/nucl.1.4.11798. Epub 2010 Mar 12. Nucleus. 2010. PMID: 21327078 Free PMC article. Review.
Cited by
- SINC, a type III secreted protein of Chlamydia psittaci, targets the inner nuclear membrane of infected cells and uninfected neighbors.
Mojica SA, Hovis KM, Frieman MB, Tran B, Hsia RC, Ravel J, Jenkins-Houk C, Wilson KL, Bavoil PM. Mojica SA, et al. Mol Biol Cell. 2015 May 15;26(10):1918-34. doi: 10.1091/mbc.E14-11-1530. Epub 2015 Mar 18. Mol Biol Cell. 2015. PMID: 25788290 Free PMC article. - MEL-28/ELYS and CENP-C coordinately control outer kinetochore assembly and meiotic chromosome-microtubule interactions.
Hattersley N, Schlientz AJ, Prevo B, Oegema K, Desai A. Hattersley N, et al. Curr Biol. 2022 Jun 6;32(11):2563-2571.e4. doi: 10.1016/j.cub.2022.04.046. Epub 2022 May 23. Curr Biol. 2022. PMID: 35609608 Free PMC article. - Probing nuclear pore complex architecture with proximity-dependent biotinylation.
Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ. Kim DI, et al. Proc Natl Acad Sci U S A. 2014 Jun 17;111(24):E2453-61. doi: 10.1073/pnas.1406459111. Epub 2014 Jun 3. Proc Natl Acad Sci U S A. 2014. PMID: 24927568 Free PMC article. - Structural and biochemical analyses of the nuclear pore complex component ELYS identify residues responsible for nucleosome binding.
Kobayashi W, Takizawa Y, Aihara M, Negishi L, Ishii H, Kurumizaka H. Kobayashi W, et al. Commun Biol. 2019 May 3;2:163. doi: 10.1038/s42003-019-0385-7. eCollection 2019. Commun Biol. 2019. PMID: 31069272 Free PMC article. - Architecture of the linker-scaffold in the nuclear pore.
Petrovic S, Samanta D, Perriches T, Bley CJ, Thierbach K, Brown B, Nie S, Mobbs GW, Stevens TA, Liu X, Tomaleri GP, Schaus L, Hoelz A. Petrovic S, et al. Science. 2022 Jun 10;376(6598):eabm9798. doi: 10.1126/science.abm9798. Epub 2022 Jun 10. Science. 2022. PMID: 35679425 Free PMC article.
References
- Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. - PubMed
- Antonin W, Franz C, Haselmann U, Antony C, Mattaj IW. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol Cell. 2005;17:83–92. - PubMed
- Beck M, Lucić V, Förster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases