GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome - PubMed (original) (raw)
GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome
Amy P Hsu et al. Blood. 2013.
Abstract
Previous reports of GATA2 mutations have focused on the coding region of the gene or full gene deletions. We recently identified 2 patients with novel insertion/deletion mutations predicted to result in mRNA nonsense-mediated decay, suggesting haploinsufficiency as the mechanism of GATA2 deficient disease. We therefore screened patients without identified exonic lesions for mutations within conserved noncoding and intronic regions. We discovered 1 patient with an intronic deletion mutation, 4 patients with point mutations within a conserved intronic element, and 3 patients with reduced or absent transcription from 1 allele. All mutations affected GATA2 transcription. Full-length cDNA analysis provided evidence for decreased expression of the mutant alleles. The intronic deletion and point mutations considerably reduced the enhancer activity of the intron 5 enhancer. Analysis of 512 immune system genes revealed similar expression profiles in all clinically affected patients and reduced GATA2 transcript levels. These mutations strongly support the haploinsufficient nature of GATA2 deficiency and identify transcriptional mechanisms and targets that lead to MonoMAC syndrome.
Figures
Figure 1
Organization and conservation within the GATA2 locus. (A) GATA2 locus. The 3 identified isoforms of human GATA2 are shown with the associated GERP, DNaseI hypersensitive sites, and reported transcription factor binding sites., (B) The conserved region within intron 5 including the composite element encompassing an E-box and GATA motifs and the ETS motif. Bold text denotes motifs for transcription factor binding. Underline denotes deletion in patient 6.II.1; *recurrent ETS motif point mutation, c.1017+572C>T. Figure modified from UCSC browser.
Figure 2
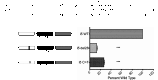
Mutation of conserved ETS motif or deletion of the composite element reduces intron 5 enhancer activity. Luciferase activity from exon 1 preceded by the wild-type intron 5 enhancer set and exon 1 preceded by the intron 5 enhancer containing either the 28 base deletion seen in patient 6.II.1 or the ETS motif C>T point mutation seen in patients 4.II.1, 11.II.1, 25.I.1, and 28.I.1. ***P < .001.
Figure 3
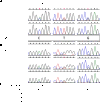
Reduced expression of mutant allele in a patient with c.1017+572C>T. (A) Genomic sequences of GATA2 exons in patient 4.II.1 and her sister 4.II.5 indicate the phase of the SNPs and the mutation. M, K, and S refer to base calls of mixed nucleotide bases at a single site, A/C, G/T, and C/G, respectively. (B) GATA2 cDNA sequences from isolated granulocytes (Grans), CD3+ T-cells (CD3+), and CD3– PBMCs (CD3–) demonstrate reduced expression of c.1017+572C>T allele. (C) Quantitation of relative peak heights of patient 4.II.1 mutant allele as a percentage of the combined peak height at that site.
Figure 4
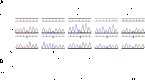
Reduced allelic expression of GATA2 in MonoMAC patients. (A) Genomic vs cDNA GATA2 sequence for patients 23.I.1, 7.I.1, 29.I.1, 28.I.1, and a healthy control. Shown is a portion of the transcript in which the patient is heterozygous at the genomic level with reduced or absent expression of 1 allele by full-length cDNA sequence. Y, S refer to base calls of mixed nucleotide bases at a single site, C/T and C/G, respectively. (B) Quantitation of relative peak heights of patient’s mutant allele as a percentage of the combined peak height at that site.
Figure 5
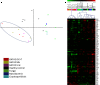
GATA2 patients have unique transcript profiles compared with healthy normals and diseased controls. Normalized transcript counts from nCounter analysis used for (A) 2-dimensional principal component analysis of patients with GATA2 i5C>T (patients 4.II.1, 11.II.1, 25.I.1), missense mutation (GATA2 Mis; patients 15.I.1, 19.II.1, 30.II.1), and reduced allelic expression without identified GATA2 mutation (GATA2 Unk; patients 7.I.1, 29.I.1) compared with healthy controls and diseased controls with similar infection types but wild-type GATA2. Fungal, disseminated coccidioidomycosis or histoplasmosis; Mycobacteria, disseminated mycobacteria; Cryptosporidiosis, disseminated cryptosporidium. (B) Hierarchical clustering of transcripts differentially regulated (P < .05) between healthy normals and GATA2 mutated patients irrespective of genotype. Samples and color labels correspond to mutations as in A.
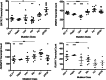
Figure 6
Transcript counts of GATA2, IKBKG, FERMT3, and RUNX1 from patients segregated by mutation class. i5C>T (n = 5, patients 4.II.1, 4.II.5, 6.II.1, 11.I.1, 25.I.1); Haplo, identified mutations resulting in loss of expression of 1 allele (n = 2, patients 13.I.2, 41.I.1); Unk, patients without identified pathogenic mutations (n = 4, patients 7.I.1, 23.I.3, 29.I.1, 33.II.2); Mis, identified GATA2 mutations predicted to result in a single amino acid change or a late frameshift with demonstrated mRNA stability (n = 8, patients 1.II.5, 5.I.1, 9.II.1, 15.I.1, 30.II.1, 33.III.3, 37.I.1, 40.I.1); compared with healthy controls (HC) (n = 9) or patients with pNTM (n = 5) with wild-type GATA2. *P < .05; **P < .01; ***P < .001.
Similar articles
- GATA2 germline mutations impair GATA2 transcription, causing haploinsufficiency: functional analysis of the p.Arg396Gln mutation.
Cortés-Lavaud X, Landecho MF, Maicas M, Urquiza L, Merino J, Moreno-Miralles I, Odero MD. Cortés-Lavaud X, et al. J Immunol. 2015 Mar 1;194(5):2190-8. doi: 10.4049/jimmunol.1401868. Epub 2015 Jan 26. J Immunol. 2015. PMID: 25624456 - Acute lymphoblastic leukemia in a patient with MonoMAC syndrome/GATA2 haploinsufficiency.
Koegel AK, Hofmann I, Moffitt K, Degar B, Duncan C, Tubman VN. Koegel AK, et al. Pediatr Blood Cancer. 2016 Oct;63(10):1844-7. doi: 10.1002/pbc.26084. Epub 2016 May 27. Pediatr Blood Cancer. 2016. PMID: 27232273 - Loss-of-function germline GATA2 mutations in patients with MDS/AML or MonoMAC syndrome and primary lymphedema reveal a key role for GATA2 in the lymphatic vasculature.
Kazenwadel J, Secker GA, Liu YJ, Rosenfeld JA, Wildin RS, Cuellar-Rodriguez J, Hsu AP, Dyack S, Fernandez CV, Chong CE, Babic M, Bardy PG, Shimamura A, Zhang MY, Walsh T, Holland SM, Hickstein DD, Horwitz MS, Hahn CN, Scott HS, Harvey NL. Kazenwadel J, et al. Blood. 2012 Feb 2;119(5):1283-91. doi: 10.1182/blood-2011-08-374363. Epub 2011 Dec 6. Blood. 2012. PMID: 22147895 Free PMC article. - MonoMAC syndrome in a patient with a GATA2 mutation: case report and review of the literature.
Camargo JF, Lobo SA, Hsu AP, Zerbe CS, Wormser GP, Holland SM. Camargo JF, et al. Clin Infect Dis. 2013 Sep;57(5):697-9. doi: 10.1093/cid/cit368. Epub 2013 May 31. Clin Infect Dis. 2013. PMID: 23728141 Free PMC article. Review. - Case Report: Missing zinc finger domains: hemophagocytic lymphohistiocytosis in a GATA2 deficiency patient triggered by non-tuberculous mycobacteriosis.
Huang X, Wu B, Wu D, Huang X, Shen M. Huang X, et al. Front Immunol. 2023 Aug 23;14:1191757. doi: 10.3389/fimmu.2023.1191757. eCollection 2023. Front Immunol. 2023. PMID: 37680631 Free PMC article. Review.
Cited by
- Oncogenic Enhancers in Leukemia.
Mulet-Lazaro R, Delwel R. Mulet-Lazaro R, et al. Blood Cancer Discov. 2024 Sep 3;5(5):303-317. doi: 10.1158/2643-3230.BCD-23-0211. Blood Cancer Discov. 2024. PMID: 39093124 Review. - Functional categorization of gene regulatory variants that cause Mendelian conditions.
Cheng YHH, Bohaczuk SC, Stergachis AB. Cheng YHH, et al. Hum Genet. 2024 Apr;143(4):559-605. doi: 10.1007/s00439-023-02639-w. Epub 2024 Mar 4. Hum Genet. 2024. PMID: 38436667 Free PMC article. Review. - Pathogenic GATA2 genetic variants utilize an obligate enhancer mechanism to distort a multilineage differentiation program.
Katsumura KR, Liu P, Kim JA, Mehta C, Bresnick EH. Katsumura KR, et al. Proc Natl Acad Sci U S A. 2024 Mar 5;121(10):e2317147121. doi: 10.1073/pnas.2317147121. Epub 2024 Feb 29. Proc Natl Acad Sci U S A. 2024. PMID: 38422019 Free PMC article. - Linking GATA2 to myeloid dysplasia and complex cytogenetics in adult myelodysplastic neoplasm and acute myeloid leukemia.
Robbins DJ, Pavletich TS, Patil AT, Pahopos D, Lasarev M, Polaki US, Gahvari ZJ, Bresnick EH, Matson DR. Robbins DJ, et al. Blood Adv. 2024 Jan 9;8(1):80-92. doi: 10.1182/bloodadvances.2023011554. Blood Adv. 2024. PMID: 38029365 Free PMC article. - From Genotype to Phenotype: How Enhancers Control Gene Expression and Cell Identity in Hematopoiesis.
Mulet-Lazaro R, Delwel R. Mulet-Lazaro R, et al. Hemasphere. 2023 Nov 8;7(11):e969. doi: 10.1097/HS9.0000000000000969. eCollection 2023 Nov. Hemasphere. 2023. PMID: 37953829 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- DK68634/DK/NIDDK NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R01 DK068634/DK/NIDDK NIH HHS/United States
- R37 DK050107/DK/NIDDK NIH HHS/United States
- R56 DK068634/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources