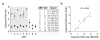Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing - PubMed (original) (raw)
doi: 10.1038/nmeth.2408. Epub 2013 Mar 17.
Abhishek Mitra, Maria Joao Silva, Magda Bienko, Norbert Dojer, Qi Wang, Elif Karaca, Roberto Chiarle, Magdalena Skrzypczak, Krzysztof Ginalski, Philippe Pasero, Maga Rowicka, Ivan Dikic
Affiliations
- PMID: 23503052
- PMCID: PMC3651036
- DOI: 10.1038/nmeth.2408
Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing
Nicola Crosetto et al. Nat Methods. 2013 Apr.
Abstract
We present a genome-wide approach to map DNA double-strand breaks (DSBs) at nucleotide resolution by a method we termed BLESS (direct in situ breaks labeling, enrichment on streptavidin and next-generation sequencing). We validated and tested BLESS using human and mouse cells and different DSBs-inducing agents and sequencing platforms. BLESS was able to detect telomere ends, Sce endonuclease-induced DSBs and complex genome-wide DSB landscapes. As a proof of principle, we characterized the genomic landscape of sensitivity to replication stress in human cells, and we identified >2,000 nonuniformly distributed aphidicolin-sensitive regions (ASRs) overrepresented in genes and enriched in satellite repeats. ASRs were also enriched in regions rearranged in human cancers, with many cancer-associated genes exhibiting high sensitivity to replication stress. Our method is suitable for genome-wide mapping of DSBs in various cells and experimental conditions, with a specificity and resolution unachievable by current techniques.
Figures
Figure 1
BLESS workflow and specificity. (a) DSBs are ligated in situ to a proximal linker (red arch) covalently linked to biotin (orange oval) (1), gDNA is extracted and fragmented (2), and labeled fragments are captured on streptavidin beads (gray ovals) (3). A distal linker (blue arch) is then ligated to the free extremity of captured fragments (4), and fragments are released by linker digestion with I-SceI (5). Released fragments are amplified by PCR using linker-specific primers (6), and sequenced (7). (b) Structure of linkers. Both proximal (P) and distal (D) linkers share an XhoI site (yellow), the I-SceI endonuclease minimal recognition site (non-highlighted letters), and a seven-thymine loop (bold). Each linker contains a specific barcode sequence marking the ligation site (orange and brown). The proximal linker is biotinylated (orange oval). (c) Proportion of fragments with proximal (P) and distal (D) barcodes in single-end (SE) and pair-end (PE) Illumina sequencing experiments. Mean ± s.d. is shown.
Figure 2
Example of HeLa breakomes associated with specific treatments. (a) Genome-wide aphidicolin (orange) and neocarzinostatin (gray) sensitivity landscapes in HeLa cells, corrected for karyotype and aphidicolin effects. Bars represent the density per one Mb bin of 48 mappable kb ASRs corrected for copy number variation effects. Individual regions and mappability maps are shown in detail in Supplementary Fig. 4b, since non-mappability can artificially lower the number of significant regions per Mb. (b) Frequency distribution of genomic distances between the centers of consecutive aphidicolin- and neocarzinostatin-sensitive regions.
Figure 3
ASRs validation. (a) Fraction of input DNA captured by ChIP in regions with significant (grey highlight) vs. non-significant aphidicolin effect in HeLa cells treated (orange) or not (green) with aphidicolin. Mean ± s.d. are shown for n = 3 biological replicates. Genomic coordinates of amplicons analyzed by qPCR are reported. Chr: chromosome. Coord: genomic coordinate. (b) Comparison of aphidicolin effect measured by BLESS vs. ChIP in regions described in (a). Captured DNA ratio: ratio of captured DNA in aphidicolin-treated (A) vs. control (C) HeLa. R: Pearson’s correlation coefficient.
Figure 4
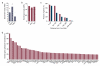
Biological characterization of ASRs. (a) Satellite repeats significantly enriched within 48 mappable kb ASRs in comparison to the rest of the genome. Repeat names follow the nomenclature in RepeatMasker . Bars: enrichments calculated based on (A1 + A2 + A3 + A4) vs. (C1 + C2 + C3 + C4) pooled samples. Diamonds: enrichments calculated based on (A1 + A2) vs. (C1 + C2) pooled samples. Dashed lines represent average genome-wide enrichment. (b) Significant enrichment of cancer-associated somatic copy number alterations. All: all alterations. Amp: amplifications. Del: deletions. Dashed lines represent average genome-wide enrichment. (c) Percentage of cancer (red) and non-cancer (blue) genes containing the center of a 48 mappable kb ASR within 2 Mb downstream from the 5′ end. (d) Ranking of aphidicolin-sensitive cancer-associated genes by decreasing sensitivity, expressed as percentage of the most sensitive gene on the left.
Similar articles
- Genome-Wide Profiling of DNA Double-Strand Breaks by the BLESS and BLISS Methods.
Mirzazadeh R, Kallas T, Bienko M, Crosetto N. Mirzazadeh R, et al. Methods Mol Biol. 2018;1672:167-194. doi: 10.1007/978-1-4939-7306-4_14. Methods Mol Biol. 2018. PMID: 29043625 - High-resolution, ultrasensitive and quantitative DNA double-strand break labeling in eukaryotic cells using i-BLESS.
Biernacka A, Skrzypczak M, Zhu Y, Pasero P, Rowicka M, Ginalski K. Biernacka A, et al. Nat Protoc. 2021 Feb;16(2):1034-1061. doi: 10.1038/s41596-020-00448-3. Epub 2020 Dec 21. Nat Protoc. 2021. PMID: 33349705 Free PMC article. - END-seq: An Unbiased, High-Resolution, and Genome-Wide Approach to Map DNA Double-Strand Breaks and Resection in Human Cells.
Wong N, John S, Nussenzweig A, Canela A. Wong N, et al. Methods Mol Biol. 2021;2153:9-31. doi: 10.1007/978-1-0716-0644-5_2. Methods Mol Biol. 2021. PMID: 32840769 - Exploring the SSBreakome: genome-wide mapping of DNA single-strand breaks by next-generation sequencing.
Zilio N, Ulrich HD. Zilio N, et al. FEBS J. 2021 Jul;288(13):3948-3961. doi: 10.1111/febs.15568. Epub 2020 Oct 2. FEBS J. 2021. PMID: 32965079 Review.
Cited by
- Target specificity of the CRISPR-Cas9 system.
Wu X, Kriz AJ, Sharp PA. Wu X, et al. Quant Biol. 2014 Jun;2(2):59-70. doi: 10.1007/s40484-014-0030-x. Quant Biol. 2014. PMID: 25722925 Free PMC article. - XPF interacts with TOP2B for R-loop processing and DNA looping on actively transcribed genes.
Chatzinikolaou G, Stratigi K, Siametis A, Goulielmaki E, Akalestou-Clocher A, Tsamardinos I, Topalis P, Austin C, Bouwman BAM, Crosetto N, Altmüller J, Garinis GA. Chatzinikolaou G, et al. Sci Adv. 2023 Nov 10;9(45):eadi2095. doi: 10.1126/sciadv.adi2095. Epub 2023 Nov 8. Sci Adv. 2023. PMID: 37939182 Free PMC article. - Innovative Precision Gene-Editing Tools in Personalized Cancer Medicine.
Dai X, Blancafort P, Wang P, Sgro A, Thompson EW, Ostrikov KK. Dai X, et al. Adv Sci (Weinh). 2020 Apr 23;7(12):1902552. doi: 10.1002/advs.201902552. eCollection 2020 Jun. Adv Sci (Weinh). 2020. PMID: 32596104 Free PMC article. Review. - Anthracyclines induce double-strand DNA breaks at active gene promoters.
Yang F, Kemp CJ, Henikoff S. Yang F, et al. Mutat Res. 2015 Mar;773:9-15. doi: 10.1016/j.mrfmmm.2015.01.007. Mutat Res. 2015. PMID: 25705119 Free PMC article. - MADDD-seq, a novel massively parallel sequencing tool for simultaneous detection of DNA damage and mutations.
Vermulst M, Paskvan SL, Chung CS, Franke K, Clegg N, Minot S, Madeoy J, Long AS, Gout JF, Bielas JH. Vermulst M, et al. Nucleic Acids Res. 2024 Sep 9;52(16):e76. doi: 10.1093/nar/gkae632. Nucleic Acids Res. 2024. PMID: 39149908 Free PMC article.
References
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nature Reviews Molecular Cell Biology. 2010;11:208–219. - PubMed
- Branzei D, Foiani M. The DNA damage response during DNA replication. Current Opinion in Cell Biology. 2005;17:568–575. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UL1TR000071/TR/NCATS NIH HHS/United States
- 250241/ERC_/European Research Council/International
- 242965/ERC_/European Research Council/International
- UL1 TR000071/TR/NCATS NIH HHS/United States
- UL1 RR029876/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials