Atomic structure and hierarchical assembly of a cross-β amyloid fibril - PubMed (original) (raw)
. 2013 Apr 2;110(14):5468-73.
doi: 10.1073/pnas.1219476110. Epub 2013 Mar 19.
Galia T Debelouchina, Marvin J Bayro, Daniel K Clare, Marc A Caporini, Vikram S Bajaj, Christopher P Jaroniec, Luchun Wang, Vladimir Ladizhansky, Shirley A Müller, Cait E MacPhee, Christopher A Waudby, Helen R Mott, Alfonso De Simone, Tuomas P J Knowles, Helen R Saibil, Michele Vendruscolo, Elena V Orlova, Robert G Griffin, Christopher M Dobson
Affiliations
- PMID: 23513222
- PMCID: PMC3619355
- DOI: 10.1073/pnas.1219476110
Atomic structure and hierarchical assembly of a cross-β amyloid fibril
Anthony W P Fitzpatrick et al. Proc Natl Acad Sci U S A. 2013.
Abstract
The cross-β amyloid form of peptides and proteins represents an archetypal and widely accessible structure consisting of ordered arrays of β-sheet filaments. These complex aggregates have remarkable chemical and physical properties, and the conversion of normally soluble functional forms of proteins into amyloid structures is linked to many debilitating human diseases, including several common forms of age-related dementia. Despite their importance, however, cross-β amyloid fibrils have proved to be recalcitrant to detailed structural analysis. By combining structural constraints from a series of experimental techniques spanning five orders of magnitude in length scale--including magic angle spinning nuclear magnetic resonance spectroscopy, X-ray fiber diffraction, cryoelectron microscopy, scanning transmission electron microscopy, and atomic force microscopy--we report the atomic-resolution (0.5 Å) structures of three amyloid polymorphs formed by an 11-residue peptide. These structures reveal the details of the packing interactions by which the constituent β-strands are assembled hierarchically into protofilaments, filaments, and mature fibrils.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
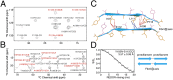
MAS NMR experiments used to probe the intermolecular arrangement of TTR(105–115) fibrils. (A and B) Shown are 2D 13C–15N ZF-TEDOR (τmix = 10.24 ms) (A) and 2D 13C–13C PDSD (τmix = 200 ms) (B) MAS NMR spectra of TTR(105–115) U–15N,13C labeled as YTIAALLSPYs and recorded at ω0H/2π = 750 MHz and ωr/2π = 12.5 kHz and ω0H/2π = 900 MHz and ωr/2π = 11 kHz, respectively, and T = 280 K. The spectra were used to constrain the intermolecular distances between the β-sheets within the TTR(105–115) protofilament. In the two spectra, intermolecular and intramolecular cross-peaks are labeled in red and black, respectively, and all cross-peaks have been assigned. (C) A cross-section of the TTR(105–115) protofilament depicting the sheet-to-sheet interface and the 11 distance constraints labeled in the spectra of A and B (red lines). The blue lines denote four additional distances between L111–13Cδ1 and –13Cδ2 and A109–15N and A108–13CO detected in additional spectra of a sample labeled as YTIAALLSPYS. The complete set of 23 intersheet distance restraints is shown in
SI Appendix, Fig. S2_B_
. Odd- and even-numbered side chains are shown as orange and violet sticks, respectively, with secondary structure shown in a cyan ribbon representation. (D) 13C–15N REDOR data obtained from a TTR(105–115) sample labeled as Y105–15N, S115–13CO2H (YTIAALLSPYS) and designed to detect interprotofilament contacts in the fibril. The intermolecular 13C–15N interaction between the terminal 15N and 13C atoms corresponds to a distance of 3.57 ± 0.06 Å, consistent with a head-to-tail protofilament arrangement as illustrated in Right. The curve was fitted by using the program SPINEVOLUTION (
SI Appendix, SI Materials and Methods
).
Fig. 2.
Atomic-resolution structure of the TTR(105–115) protofilament determined by MAS NMR. The structures (calculated with CNSsolve; see
SI Appendix, SI Materials and Methods
) have a rmsd of ∼0.4 Å for the backbone and 0.7 Å for all atoms. Odd- and even-numbered side chains are shown as orange and violet sticks, respectively, with secondary structure shown in a cyan ribbon representation. (A) The β-sheet viewed perpendicularly to the fibril axis illustrating the parallel in-register β-strands and the hydrogen bonds defining the β-sheet (yellow lines). The conformation was determined from eight 13C=O–13C=O double quantum distance measurements and one 13C=O–13C=O REDOR distance measurement (
SI Appendix, Table S1
). (B) Cross-sectional view of the two-sheet protofilament along the peptide chain direction. There is a sheet–sheet offset of approximately one-fifth the separation of hydrogen-bonded β-strands (i.e., 0.2 × 4.67 Å = 0.93 Å) shown clearly by the interdigitation of the Y105 (orange sticks) and Y114 (violet sticks) side chains. (C) Protofilament–protofilament interface viewed looking down the long axis of the fibril showing the head-to-tail packing arrangement. Interprotofilament hydrogen bonds between the terminal C=O and N–H groups and between the Y105 OH atoms and the S115 Oγ atoms are depicted as yellow dashes.
Fig. 3.
Representative cryo-EM images of averaged fibrils (class averages), surface representations of reconstructions, 2D projections, and contoured density cross-sections of the three types of fibril formed by TTR(105–115). (A) Doublet class average (Left), 3D reconstruction (Center; orange), and 2D projection of the fibril reconstruction (Right). (B) Triplet class average (Left), 3D reconstruction (Center; yellow), and 2D projection of the fibril reconstruction (Right). (C) Quadruplet class average (Left), 3D reconstruction (Center; purple), and 2D projection of the fibril reconstruction (Right). (D_–_F) Contoured density cross-sections of the doublet (D), triplet (E), and quadruplet (F) fibrils. Contours represent density levels of multiples of 0.5σ above the average density of the background (outermost contour). Measurements of dimensions were determined at 1.0σ above the mean density.
Fig. 4.
Atomic-resolution cross-sections of the three types of TTR(105–115) amyloid fibrils determined by combining MAS NMR and cryo-EM. Reconstructed cross-section density maps are shown as two electron density isosurfaces (1.0σ and 2.2σ above the mean density) with the secondary structure of the constituent NMR-derived protofilaments shown in a cyan ribbon representation. (A–C) The doublet (A), triplet (B), and quadruplet (C) fibril cross-sections can accommodate pairs of two, three, and four interconnected protofilaments, respectively. (D_–_F) All-atom representation of the doublet (D), triplet (E), and quadruplet (F) cross-sections with cryo-EM envelopes superimposed. Cryo-EM envelopes are shown as orange, yellow, and purple contours at 1.0σ above the mean density for doublet, triplet, and quadruplet fibrils, respectively. For a discussion on the resolution of the cryo-EM maps, see
SI Appendix, Table S4
.
Fig. 5.
Five diverse biophysical techniques were integrated to determine unambiguously the structures of each of the motifs that make up the TTR(105–115) amyloid fibrils. Spanning five orders of magnitude, the overlapping length scales of MAS NMR (0.1–10 Å), X-ray diffraction (3–100 Å), cryo-EM (8–1,000 Å), AFM (30–1,000 Å), and STEM (80–1,000 Å) enabled us to derive self-consistent, high-precision structural restraints on the secondary (β-strand and -sheet; distance restraints of <6 Å), tertiary (protofilament; distance restraints of 4.5–37 Å), and quaternary structure (filament and fibril; distance restraints of 16–1,000 Å) of the TTR(105–115) amyloid fibrils. (A) Histogram of STEM MPL measurements of TTR(105–115) fibrils, which reveals three populations of fibrils, with a best fit (gray solid line) being the sum of three Gaussian curves with values of 2.5 ± 0.3 kDa/Å (orange solid line), 3.3 ± 0.3 kDa/Å (yellow solid line), and 4.1 ± 0.3 kDa/Å (purple solid line). The orange, yellow, and purple dashed lines refer to the number of TTR(105–115) peptides per 4.67-Å repeat in the doublet (8 peptides), triplet (12 peptides), and quadruplet (16 peptides) fibrils, respectively. (B) Comparison of the high-resolution experimental X-ray diffraction pattern from TTR(105–115) fibrils (34) (Left) and the simulated X-ray diffraction pattern for TTR(105–115) fibrils (Right). The fibril axis is vertical, with the incident beam directed orthogonally to this axis. The meridional reflection at 4.67 Å and the equatorial reflection at 8.86 Å are characteristic of cross-β structure. (C) High-resolution AFM image of fibrils (pink and purple) and filaments (green) formed by TTR(105–115). (Scale bar, 1 μm.) Fibrils (pink and purple) have heights ranging from 70 to 160 Å and pitches of 950 ± 100 Å. The filament has an average height of 38.7 ± 4.4 Å. (D) Hierarchy of atomic-resolution motifs involved in the self-assembly of the amyloid fibrils and their polymorphism.
Fig. 6.
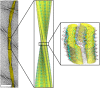
Close-up view of the MAS NMR atomic-resolution structure of the triplet fibril fitted into the cryo-EM reconstruction (Center). The background image of the fibril (Left) was taken using TEM. (Scale bar, 50 nm.) The fibril surfaces (Right) are shown at 1.0σ (white) and 2.2σ (yellow) above the mean density, respectively, and the constituent β-sheets are shown in a ribbon representation; oxygen, carbon, and nitrogen atoms are shown in red, gray, and blue, respectively.
Similar articles
- The architecture of amyloid-like peptide fibrils revealed by X-ray scattering, diffraction and electron microscopy.
Langkilde AE, Morris KL, Serpell LC, Svergun DI, Vestergaard B. Langkilde AE, et al. Acta Crystallogr D Biol Crystallogr. 2015 Apr;71(Pt 4):882-95. doi: 10.1107/S1399004715001674. Epub 2015 Mar 27. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 25849399 Free PMC article. - Accurate determination of interstrand distances and alignment in amyloid fibrils by magic angle spinning NMR.
Caporini MA, Bajaj VS, Veshtort M, Fitzpatrick A, MacPhee CE, Vendruscolo M, Dobson CM, Griffin RG. Caporini MA, et al. J Phys Chem B. 2010 Oct 28;114(42):13555-61. doi: 10.1021/jp106675h. J Phys Chem B. 2010. PMID: 20925357 Free PMC article. - Atomic models of de novo designed cc beta-Met amyloid-like fibrils.
Steinmetz MO, Gattin Z, Verel R, Ciani B, Stromer T, Green JM, Tittmann P, Schulze-Briese C, Gross H, van Gunsteren WF, Meier BH, Serpell LC, Müller SA, Kammerer RA. Steinmetz MO, et al. J Mol Biol. 2008 Feb 22;376(3):898-912. doi: 10.1016/j.jmb.2007.11.100. Epub 2007 Dec 5. J Mol Biol. 2008. PMID: 18178219 - Amyloid structure and assembly: insights from scanning transmission electron microscopy.
Goldsbury C, Baxa U, Simon MN, Steven AC, Engel A, Wall JS, Aebi U, Müller SA. Goldsbury C, et al. J Struct Biol. 2011 Jan;173(1):1-13. doi: 10.1016/j.jsb.2010.09.018. Epub 2010 Sep 22. J Struct Biol. 2011. PMID: 20868754 Free PMC article. Review. - Molecular structures of amyloid and prion fibrils: consensus versus controversy.
Tycko R, Wickner RB. Tycko R, et al. Acc Chem Res. 2013 Jul 16;46(7):1487-96. doi: 10.1021/ar300282r. Epub 2013 Jan 7. Acc Chem Res. 2013. PMID: 23294335 Free PMC article. Review.
Cited by
- A Structural Model for a Self-Assembled Nanotube Provides Insight into Its Exciton Dynamics.
Gao M, Paul S, Schwieters CD, You ZQ, Shao H, Herbert JM, Parquette JR, Jaroniec CP. Gao M, et al. J Phys Chem C Nanomater Interfaces. 2015 Jun 18;119(24):13948-13956. doi: 10.1021/acs.jpcc.5b03398. Epub 2015 May 26. J Phys Chem C Nanomater Interfaces. 2015. PMID: 26120375 Free PMC article. - Multi-_e_GO: Model Improvements toward the Study of Complex Self-Assembly Processes.
Bačić Toplek F, Scalone E, Stegani B, Paissoni C, Capelli R, Camilloni C. Bačić Toplek F, et al. J Chem Theory Comput. 2024 Jan 9;20(1):459-468. doi: 10.1021/acs.jctc.3c01182. Epub 2023 Dec 28. J Chem Theory Comput. 2024. PMID: 38153340 Free PMC article. - Differential effects of glycation on protein aggregation and amyloid formation.
Iannuzzi C, Irace G, Sirangelo I. Iannuzzi C, et al. Front Mol Biosci. 2014 Sep 2;1:9. doi: 10.3389/fmolb.2014.00009. eCollection 2014. Front Mol Biosci. 2014. PMID: 25988150 Free PMC article. Review. - Structure of cytotoxic amyloid oligomers generated during disaggregation.
Kaku T, Ikebukuro K, Tsukakoshi K. Kaku T, et al. J Biochem. 2024 May 31;175(6):575-585. doi: 10.1093/jb/mvae023. J Biochem. 2024. PMID: 38430131 Free PMC article. Review. - Biochemical Principles in Prion-Based Inheritance.
Dennis EM, Garcia DM. Dennis EM, et al. Epigenomes. 2022 Jan 25;6(1):4. doi: 10.3390/epigenomes6010004. Epigenomes. 2022. PMID: 35225957 Free PMC article. Review.
References
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. - PubMed
- Sunde M, et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273(3):729–739. - PubMed
- Knowles TPJ, Smith JF, Craig A, Dobson CM, Welland ME. Spatial persistence of angular correlations in amyloid fibrils. Phys Rev Lett. 2006;96(23):238301. - PubMed
- Knowles TP, et al. Role of intermolecular forces in defining material properties of protein nanofibrils. Science. 2007;318(5858):1900–1903. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0200653/MRC_/Medical Research Council/United Kingdom
- P41 EB002026/EB/NIBIB NIH HHS/United States
- EB-003151/EB/NIBIB NIH HHS/United States
- 089703/WT_/Wellcome Trust/United Kingdom
- EB-002026/EB/NIBIB NIH HHS/United States
- R01 EB003151/EB/NIBIB NIH HHS/United States
- BB/C00759X/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources