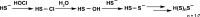Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery - PubMed (original) (raw)
Review
. 2013 Jul 10;113(7):4633-79.
doi: 10.1021/cr300163e. Epub 2013 Mar 20.
Affiliations
- PMID: 23514336
- PMCID: PMC4303468
- DOI: 10.1021/cr300163e
Free PMC article
Review
Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery
Candice E Paulsen et al. Chem Rev. 2013.
Free PMC article
No abstract available
Figures
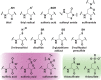
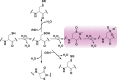
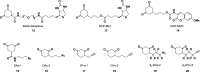
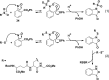
Chart 1. Biologically Relevant Cysteine Chemotypesa
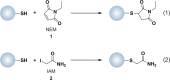
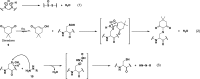
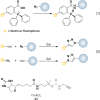
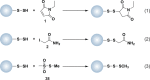
Chart 2. Protein Thiols React with _N_-Ethylmaleimide (NEM, 1, Equation 1) and Iodoacetamide (IAM, 2, Equation 2) by Michael Addition or SN2 Displacement, Respectively
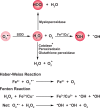
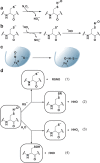
Chart 3. Formation and Transformation of Biologically Relevant Reactive Oxygen Species (ROS)a
Figure 1
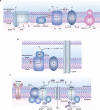
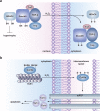
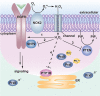
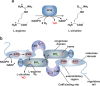
Biological sources of reactive oxygen species (ROS). (a) The mitochondrial electron transport chain (ETC). Four protein complexes (I–IV) funnel electrons (black arrows) from NADH and succinate in the matrix to ultimately reduce molecular oxygen to water and establish a proton gradient (gray arrows) that is harnessed by complex V to generate ATP. Electrons can leak prematurely from the ETC at complexes I and III (red arrows) to generate superoxide (O2•–) in either the matrix or intermembrane space. (b) p66 (Shc) facilitates pro-apoptotic O2•– or H2O2 production in the mitochondria. In response to UV irradiation or growth factor deprivation, p66 (Shc) localizes to the mitochondria where it interacts with complex III to divert electrons from cytochrome c directly to molecular oxygen to generate O2•– or H2O2. This H2O2 can translocate to the cytoplasm (not shown) where it can influence signaling, and can regulate opening of the mitochondrial permeability transition pore (mPTP), which initiates mitochondrial swelling and apoptosis. (c) NOX enzyme complexes assemble at distinct regions of the plasma membrane or intracellular membranes to regulate localized ROS production in response to diverse signals. Receptor stimulation initiates the recruitment of specific coactivating proteins or calcium to one of seven NOX catalytic cores. Once activated, NOX enzymes funnel electrons from NADPH in the cytoplasm through FAD and heme cofactors across the membrane to generate O2•– (NOX1-2) or H2O2 (Duox1-2) on the extracellular/lumenal face. O2•– is dismutated to H2O2 and oxygen either spontaneously or as enhanced by SOD, which can translocate across the membrane by diffusion or, more likely, through aquaporin channels to regulate protein activity and signaling in the cytoplasm.
Figure 2
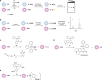
Oxidative modification of cysteine residues by hydrogen peroxide (H2O2). The initial reaction product of a low p_K_a protein thiolate with H2O2 yields a sulfenic acid, whose stability is determined, in part, by its accessibility to additional thiols. Reaction with a second cysteine in the same or neighboring protein yields a disulfide. Alternatively, reaction with the low molecular weight thiol, glutathione (GSH) affords a specialized mixed disulfide called a glutathione disulfide. In some proteins in which a neighboring cysteine is not present, nucleophilic attack of a backbone amide on the sulfenic acid yields a cyclic sulfenyl amide. Each of these oxoforms can be reduced by the GSH/glutaredoxin or thioredoxin/thioredoxin reductase systems to regenerate the reduced thiolate (not shown). In the presence of excess H2O2, such as under conditions of oxidative stress, the sulfenic acid can be hyperoxidized to the largely irreversible sulfinic and sulfonic acid forms (red box).
Figure 3
General overview of indirect and direct chemical methods to study protein oxidation. (a) Loss of labeling of oxidized thiols by an alkylating agent indirectly monitors protein oxidation. In response to oxidant treatment, susceptible cysteines are oxidized (purple) and thus are less reactive with alkylating agents such as NEM or IAM. Use of a biotinylated or fluorophore-conjugated alkylating agent permits detection by avidin blot or in-gel fluorescence, in which oxidized proteins exhibit a loss of signal. (b) Differential alkylation of reduced and oxidized thiols indirectly monitors protein oxidation. Free thiols (blue) are blocked with an alkylating agent such as NEM or IAM, reversibly oxidized thiols (purple) are reduced with a reducing agent such as DTT or TCEP, and nascent thiols are labeled with a second alkylating agent conjugated to biotin or a fluorophore. Oxidized proteins exhibit enhanced signal by avidin blot or in-gel fluorescence. (c) Direct chemical method to detect specific cysteine oxoforms. Samples are treated with a biotin or fluorophore-conjugated probe that selectively reacts with a distinct cysteine chemotype (purple) in which signal by avidin blot or in-gel fluorescence increases with increased protein oxidation.
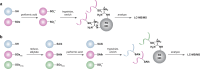
Figure 4
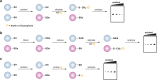
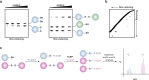
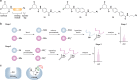
Indirect chemical methods to study general cysteine oxidation. (a) Loss of labeling of oxidized thiols by biotinylated-iodoacetamide (BIAM) indirectly monitors protein oxidation. Oxidized cysteines (purple) exhibit decreased reactivity with BIAM than reduced thiols, and are observed as a loss of signal by avidin blot. (b) Isotope-coded affinity tag (ICAT) reagents determine the ratio of oxidized thiols. Samples are untreated or subjected to oxidant. Free thiols are subsequently labeled with a light (12C) ICAT reagent in the untreated sample and with a heavy (13C) ICAT reagent in the oxidant-treated sample. As in panel a, reactive thiols (purple) exhibit decreased labeling upon oxidation. The samples are mixed, trypsinized, and enriched via the biotin affinity tag on the ICAT reagent. Eluted peptides are analyzed by LC-MS and heavy and light ICAT-labeled peptides are chemically identical, but differ in mass by 9 Da. The fraction of a thiol oxidized in the sample is determined by the ratio of heavy (13C) to light (12C) signal intensity, whereby thiols that are susceptible to oxidation (purple) will exhibit decreased signal intensity with the heavy ICAT reagent.
Figure 5
Possible fates of protein disulfides. Once formed, a protein disulfide (inter- or intramolecular) can undergo thiol-disulfide exchange with a third cysteine within the same or neighboring protein (eq 1). Herein, p_K_a of the disulfide thiols and thiol accessibility influence which cysteine is expelled. In the presence of high concentrations of H2O2, disulfides can additionally be oxidized to the thiosulfinate and thiosulfonate forms, though these reactions are very slow. Because of the potential for resonance stabilization or decreased p_K_a, subsequent reaction of these intermediates with a third cysteine affords a disulfide and a sulfenic acid (eq 2) or sulfinic acid (eq 3). The biological relevance of the thiosulfinate and thiosulfonate modifications is unknown due to a lack of means to study these oxoforms, however, a thiosulfinate forms as an intermediate during the sulfiredoxin catalytic cycle.
Figure 6
Disulfide-mediated redox regulation of subcellular localization and protein–protein interactions. (a) Model for redox regulation of cardiac hypertrophy by HDAC4. The class II histone deacetylase HDAC4 normally deacetylates histones to suppress expression of genes involved in cardiac hypertrophy. Nuclear localization of HDAC4 is mediated by its association with importin α (Imp) via a multiprotein complex including the small molecular chaperone DnaJb5, the thioredoxin binding protein TBP-2, and thioredoxin (Trx1). In response to oxidant, HDAC4 and DnaJb5 undergo intramolecular disulfide bond formation, which causes dissociation and nuclear export of the complex permitting derepression of genes involved in hypertrophy. Upon removal of H2O2, Trx1 is believed to reduce the disulfides in HDAC4 and DnaJb5 to restore assembly and nuclear localization of the complex (not shown). (b) Model for redox regulation of apoptosis by cofilin. Cofilin associates with actin in the cytoplasm to disassemble actin filaments for cytoskeletal reorganization. In the presence of H2O2, two intramolecular disulfides form in cofilin permitting its relocation to the mitochondria by an unresolved mechanism. In the mitochondria, cofilin interacts with the mPTP to stimulate pore opening, mitochondrial swelling, cytochrome c release, and ultimately induction of apoptosis.
Figure 7
Methods for detection and identification of protein disulfides. (a) Differential migration of proteins containing intra- and intermolecular disulfide bonds. Samples are resolved under nonreducing SDS-PAGE conditions. Intramolecular disulfides can facilitate enhanced protein migration in some proteins as compared to the reduced species (left). Intermolecular disulfide complexes migrate at the combined molecular weight of the individual proteins (right). (b) Redox 2D-PAGE. Protein samples are first separated by nonreducing gel electrophoresis to separate disulfide-bonded complexes by size (top). The proteins are subsequently reduced in-gel with DTT, alkylated with NEM or IAM, and separated in the second dimension under reducing conditions (down). Proteins that are not involved in intermolecular disulfide complexes run at the diagonal. Proteins involved in disulfide complexes migrate off the diagonal and can be identified by in-gel digestion and LC-MS/MS (not shown). (c) OxICAT method combines the ICAT technology with differential alkylation of reduced and oxidized thiols to permit quantification of oxidized residues. Cell lysates are generated in the presence of trichloroacetic acid and detergents to facilitate exposure of all protein cysteines while inhibiting thiol/disulfide exchange. Reduced thiols (blue) are subsequently blocked with the light (12C) ICAT reagent (blue), oxidized proteins (purple) are reduced with TCEP, and nascent thiols are alkylated with the heavy (13C) ICAT reagent (purple). Samples are trypsinized and labeled peptides are avidin enriched. Eluted peptides are analyzed by LC-MS and heavy and light ICAT-labeled peptides are chemically identical, but differ in mass by 9 Da. The percentage of a particular thiol that is oxidized in a sample is determined by the ratio of heavy (13C) to light (12C) signal intensity from the corresponding peptide. While TCEP can reduce all reversible oxoforms (e.g., disulfides, sulfenic acid, _S_-nitrosothiols), sulfenic acids and _S_-nitrosothiols are often acid-labile and likely lost during sample preparation. As such, OxICAT is likely most suitable to detect cysteines involved in disulfide bonds.
Figure 8
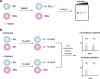
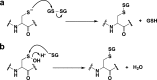
Mechanisms for glutathionylation. Protein glutathionylation products can be formed by (a) thiol/disulfide exchange of a protein thiolate with oxidized glutathione (GSSG) or (b) condensation of GSH with a protein sulfenic acid.
Figure 9
Methods to detect protein S-glutathionylation. (a) GST overlay. Samples are separated by nonreducing SDS-PAGE to preserve protein-GSH disulfides. Blots are subsequently treated with biotinylated glutathione S-transferase (GST), which binds selectively to GSH and permits detection of S-glutathionylated proteins by avidin blot. (b) Indirect differential alkylation of S-glutathionylated proteins. Free thiols are blocked with NEM or IAM, protein-GSH disulfides are selectively reduced with glutaredoxin (Grx), and nascent thiols are labeled with an alkylating agent conjugated to biotin or a fluorophore. S-Glutathionylated proteins are detected by avidin blot or in-gel fluorescence. (c) Biotinylated glutathione ethyl ester (BioGEE, 3) enables in situ detection of glutathionylated proteins. N,_N_-Biotinyl glutathione disulfide (4) permits detection of proteins that become S-glutathionylated by thiol/disulfide exchange with GSSG.
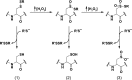
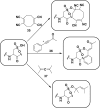
Chart 4. Sulfenic Acids Exhibit Both Nucleophilic and Electrophilic Character, As Illustrated by Condensation of Two Sulfenic Acids to Afford a Thiosulfinate (Black Box)a
Chart 5. Reaction Schemes of Condensation of Two Sulfenic Acids to Yield a Thiosulfinate (Equation 1) and Electrophilic Reaction of Sulfenic Acid with Dimedone (9, Equation 2) and Hydrazines (10, Equation 3)
Figure 10
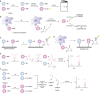
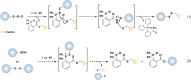
Methods to detect protein sulfenic acids. (a) Indirect differential alkylation of protein sulfenic acids. Free thiols (blue) are blocked with NEM or IAM, protein sulfenic acids (purple) are reduced with arsenite, and nascent thiols are labeled with biotinylated NEM (NEM-Biotin). Sulfenylated proteins are detected by avidin blot where increased protein oxidation is observed as an increase in signal intensity. (b) Direct in situ labeling of protein sulfenylation. Cells are treated with or without stimulant (e.g., oxidant, growth factor) and subsequently incubated with azido or alkyne dimedone analogues, such as 18 to chemically modify sulfenylated proteins. Afterward, excess probe is removed, cell lysates are generated, and probe-modified proteins are conjugated to biotin or a fluorophore by a coupling reaction (e.g., Staudinger ligation or Huisgen [3 + 2] cycloaddition). The samples can then be avidin enriched and subjected to proteomics analysis or analyzed by avidin blot or in-gel fluorescence where increased protein sulfenylation correlates to enhanced signal intensity. (c and d) High-throughput immunological detection of dimedone (9)-modified proteins using arrays. (c) Proteins immobilized on a microarray that are susceptible to sulfenylation are irreversibly modified by 9. The protein–dimedone adduct forms an epitope for selective detection by the antibody. (d) Cells are treated with or without stimulant (e.g., oxidant, growth factor) and are subsequently incubated with 9 to irreversibly modify sulfenylated proteins. Subsequent to cell lysis, proteins within a given signaling pathway are immobilized on an antibody array and dimedone-modified proteins are detected by addition of the antibody. (e) Isotope-coded dimedone 2-iododimedone (ICDID) permits quantification of protein sulfenylation. Sulfenic acids are labeled by _d_6-dimedone (21, purple), then excess reagent is removed and free thiols are labeled by _d_0-2-iododimedone (22, blue) generating chemically identical adducts that differ by 6 Da. The samples are trypsinized and analyzed by LC-MS where the extent of sulfenic acid occupancy is determined by the ratio of _d_6-dimedone to _d_0-dimedone peak intensities. (f) Quantification and site-identification of protein sulfenic acids with _d_6-DAz-2 or _d_6-DYn-2 and an acid-cleavable linker (ACL) coupling reagent. Sulfenic acids are labeled with d_0-DAz-2 (16) in the untreated sample and with d_6-DAz-2 (19) in the oxidant-treated sample. Excess probe is removed and the samples are combined and biotinylated by coupling with the alkyne-ACL (23) to generate chemically identical adducts that differ by 6 Da. The sample is then trypsinized, avidin enriched, and trifluoroacetic acid-eluted peptides are analyzed by LC-MS/MS where the increase in sulfenic acid modification in response to oxidant is determined by the ratio of d_6 to d_0 peak intensities. Biotin can complicate spectra and decreases peptide recovery, and the removal of biotin with the ACL permits increased sample elution and direct identification of modified peptides in the MS.
Chart 6. Biotin, Fluorophore, and Chemical Handle Derivatives of Dimedone
Chart 7. Bioorthogonal Coupling Reactions Staudinger Ligation and Huisgen [3 + 2] Cycloadditiona
Figure 11
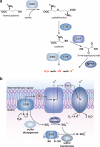
Redox regulation of epidermal growth factor (EGF) signaling by protein sulfenylation. Binding of EGF to the EGF receptor (EGFR) facilitates receptor dimerization, activation (not shown), and promotion of NOX2 complex assembly. NOX2-derived H2O2 translocates into the cytoplasm likely through channels such as aquaporins where it has been shown to regulate the activity of proteins involved in the EGFR signaling cascade. EGFR and the phosphatases SHP2, PTEN, and PTP1B were all found to be sulfenylated in response to EGF stimulation, albeit with differential sensitivities (ranked 1–4 in order of decreasing susceptibility). The sensitivity of each protein to oxidation correlates to their relative proximity to the oxidant source. In this way, EGFR, which forms a complex with NOX2, exhibited the highest sensitivity. Moreover, the EGFR-associated phosphatase SHP2 exhibited increased susceptibility to sulfenylation as compared to the cytoplasmic phosphatase PTEN, which regulates the levels of PIP3, and PTP1B, which is localized to the cytoplasmic face of the endoplasmic reticulum (ER). Co-localization of antioxidant enzymes such as peroxiredoxins (Prx) to the signaling regions is thought to limit the range of H2O2 diffusion (green area). Interestingly, NOX-derived reactive oxygen species (ROS) have also been shown to inactivate Prxs by hyperoxidation (PrxII) or phosphorylation (PrxI). These regulatory mechanisms have been proposed to permit localized accumulation of ROS for redox regulation of proteins located near the oxidant source (pink area).
Figure 12
Targeted approaches to monitor cysteine oxPTM with in specific or family of proteins. (a) Redox-based probes (RBPs, 24–26) for protein tyrosine phosphatases (PTPs) are comprised of three parts: a warhead that permits chemoselective reaction with sulfenic acid, a PTP-directed scaffold that exhibits enhanced affinity for target binding, and an azide chemical reporter to facilitate downstream detection and isolation of labeled protein. (b) Indirect two-stage immunochemical approach to monitor PTP oxidation. In stage 1, free thiols in one aliquot of sample are alkylated with NEM, oxidized cysteines are reduced with DTT, and nascent thiols are hyperoxidized to sulfonic acid with pervanadate. The proteins are subsequently trypsinized, and sulfonic acid-modified peptides are isolated with a monoclonal antibody that recognizes hyperoxidized PTPs. In stage 2, a second aliquot of sample is reduced with DTT and all thiols are oxidized to sulfonic acid with pervanadate and processed as in stage 1. The enriched peptides are analyzed by LC-MS/MS and the extent of PTP oxidation is determined by the ratio of stage 1 to stage 2 signal intensities. (c) Conformation-sensing single-chain variable antibodies that selectively recognize the unique conformation of the sulfenyl amide oxoform of PTP1B.
Chart 8. Resonance Structures of Sulfinic Acids
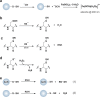
Chart 9. Sulfinic Acids Function as Soft Nucleophiles Reacting Primarily from the Sulfur to Undergo Alkylation (27) or Nucleophilic Addition to Activated Alkenes (28), Aldehydes (29), Lactones (30), α,β-Unsaturated Compounds (31), and Diazonium Salts (32)a
Chart 10. Chemoselective Approach to Detect Sulfinic Acidsa
Chart 11. Reactions of Sulfonic Acidsa
Figure 13
Method for selective enrichment of sulfonic acid-modified peptides. (a) All cysteine residues are oxidized to sulfonic acid with performic acid. Proteins are then trypsinized and sulfonic acid-modified peptides are enriched using polyarginine (PA)-coated nanodiamonds (ND). Eluted peptides are analyzed by LC-MS/MS. (b) A plausible extension of the PA-ND enrichment technology to identify sulfinylated and sulfonylated cysteines. Samples are first treated with a reducing agent to reduce all reversibly oxidized cysteines (purple), and alkylated with NEM or IAM. Irreversibly oxidized cysteines (green) are subsequently oxidized to sulfonic acid with performic acid. The sample is then trypsinized, sulfonic acid-modified peptides are enriched with PA-ND, and eluted peptides are analyzed by LC-MS/MS to identify sites of hyperoxidation.
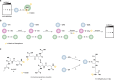
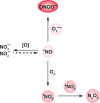
Chart 12. Formation and Transformation of Biologically Relevant Reactive Nitrogen Species (RNS)a
Figure 14
Nitric oxide (•NO) production by nitric oxide synthases (NOS). (a) Reaction catalyzed by NOS. (b) Linear arrangement of NOS. NOS contain three distinct domains: the N-terminal oxygenase domain (gray), the C-terminal reductase domain (light blue), and the connecting calmodulin (CaM) binding site (purple). All three NOS isoforms encode a C-terminal regulatory tail and endothelial NOS (eNOS) and neuronal NOS (nNOS) additionally contain an autoinhibitory region in the reductase domain. Activation of eNOS and nNOS requires binding of Ca2+/CaM whereas inducible NOS (iNOS) is expressed with Ca2+/CaM tightly bound. The production of •NO by NOS involves translocation of electrons from NADPH bound in the reductase domain through the FAD and FMN cofactors. The electrons are then transferred to the heme prosthetic group in the oxygenase domain where
l
-arginine and molecular oxygen bind. The tetrahydrobiopterin (BH4) cofactor in the oxygenase domain appears to regulate the nature of reactive intermediates generated by NOS (e.g., •NO versus O2•–). The CaM binding site, autoinhibitory region, and regulatory tail are believed to regulate enzyme activity by influencing the efficiency of electron transfer between the reductase and oxygenase domains. All NOS isoforms function as homodimers that are stabilized by a zinc ion coordinated by two cysteine residues from each monomer and there is evidence suggesting that electrons are transferred between monomers (as depicted). NOS enzymes additionally contain sequences such as PDZ domains (deep blue) that facilitate protein–protein interactions, which are involved in subcellular targeting of NOS and for mediating direct interactions with protein targets of •NO.
Figure 15
Formation and subsequent reactions of _S_-nitrosothiols. Three prominent mechanisms for _S_-nitrosothiol formation include (a) reaction of a protein or low molecular weight thiolate with N2O3, (b) formation of a thiyl radical upon initial reaction of a thiolate with •NO2 and other radical species and subsequent radical–radical combination with •NO, and (c) autotransfer of heme-bound +NO to a nearby cysteine thiolate as has been demonstrated for hemoglobin and nitrophorin. (d) Once formed, an _S_-nitrosothiol can react with a neighboring cysteine residue either within the same or an adjacent protein, or with GSH (not shown) to undergo transnitrosylation (eq 1) or disulfide bond formation (eq 2). Alternatively, an _S_-nitrosothiol can be hydrolyzed to release the free thiolate and nitrite (HNO2) (eq 3) or a sulfenic acid and HNO (eq 4). In each case, the p_K_a of the sulfur in the _S_-nitrosothiol, in part, influences which product is formed. In most cases, transnitrosylation and release of a free thiolate are favored upon reaction with a second cysteine or water, respectively due to the high p_K_a of the HNO leaving group.
Figure 16
Regulation of neuronal signaling by S-nitrosylation. PSD-95 is a scaffolding protein that localizes to postsynaptic densities by reversible S-palmitoylation of two cysteine residues in the N-terminal region. In the absence of ligand, nNOS is physically linked to the NMDA receptor (NMDAR) at the neuronal plasma membrane via a mutual interaction with PSD-95. PSD-95 similarly localizes stargazin near NMDAR. NMDA binding to NMDAR facilitates calcium entry, which activates nNOS mediating •NO production and subsequent S-nitrosylation of PSD-95 on the same cysteine residues that undergo S-palmitoylation. S-nitrosylation thereby prevents PSD-95 lipidation and decreases membrane association of PSD-95 and, hence, nNOS. Stargazin is similarly S-nitrosylated, which enhances its interaction with the AMPA receptor facilitating its recruitment to the postsynaptic densities.
Chart 13. Predicted Mechanism for the Reaction of Ascorbate with _S_-Nitrosothiol
Figure 17
Indirect and direct chemical methods for _S_-nitrosothiol detection. (a) The biotin switch technique (BST) is an indirect differential alkylation method that involves blocking free thiols (blue) with methylmethane thiosulfonate (MMTS, 38), reducing _S_-nitrosothiols (purple) with ascorbate, and labeling nascent thiols with Biotin-HPDP. The samples are then analyzed by nonreducing avidin blot in which _S_-nitrosylation correlates to increased signal intensity. (b) Quantification of protein S-nitrosylation with d-Switch. The d-Switch technique combines the BST with isotopically labeled NEM in which free thiols (blue) are blocked with _d_0-NEM, _S_-nitrosothiols (purple) are reduced with ascorbate and labeled with _d_5-NEM. The samples are subsequently separated by SDS-PAGE, digested in-gel with trypsin, and the resulting peptides are analyzed by LC-MS/MS. The extent of protein S-nitrosylation is determined by the ratio of _d_5-NEM to _d_0-NEM signal intensity. (c–e) Triarylphosphine-based methods to directly modify _S_-nitrosothiols. (c) Triarylphosphine reagent 40 reacts with _S_-nitrosothiols to yield a disulfide-bonded biotin adduct. (d) Compound 41 is oxidized upon reaction with _S_-nitrosothiols to yield a fluorescent compound that comments on the presence of _S_-nitrosothiols, but does not covalently modify oxidized proteins. (e) Water-soluble triarylphosphine 42 appears to form a stable _S_-alkylphosphonium adduct as monitored by 31P NMR and mass spectrometry.
Chart 14. Reaction Mechanism of Triarylphosphine 40 with a Protein _S_-Nitrosothiol to Yield a Disulfide-Bonded Biotin Adduct (Equation 1) and Potential Reaction of Sulfenic Acids or Disulfides with 40 (Dashed Arrows) Should Not Yield the Same Adduct (Equation 2)
Chart 15. Formation of H2S2 and Higher Order Polysulfides by Reaction of Hydrogen Sulfide (H2S) with ROS, Such as Hypochlorous Acid (HOCl) Initially Yields a Sulfenyl Chloride That Is Hydrolyzed by Water to Afford a Sulfenic Acid
Figure 18
Generation and metabolism of H2S. (a) Pyridoxal 5′-phosphate (PLP)-dependent enzymes of the transsulfuration pathway, cystathionine β-synthase (CBS) and cystathionine γ-lyase (CSE) catalyze H2S production from homocysteine, cystathionine, and cysteine. Additionally, the combined activity of cysteine aminotransferase (CAT) and 3-mercaptopyruvate sulfurtransferase (3MST) in the cytoplasm and mitochondria can generate H2S. (b) Oxidative metabolism of H2S in the mitochondrial matrix is catalyzed by a series of enzymes to generate persulfide, sulfite (SO3–), thiosulfate (S2O32—), and sulfate (SO42—). The first step is catalyzed by SQR, which forms a protein-bound persulfide intermediate and funnels electrons from H2S oxidation directly into the ETC. The subsequent action of sulfur dioxygenase, sulfite oxidase and sulfur transferase are proposed to convert SQR persulfide into SO3–, SO42–, and S2O32–, respectively.
Chart 16. Colorimetric and Fluorescent Methods for Detection and Quantification of H2Sa
Figure 19
Formation and reactions _S_-sulfhydryls. (a) The cyanolysis assay for persulfides relies upon reaction of persulfides with cyanide (CN–) to form thiocyanate (SCN–). SCN– is subsequently detected with iron nitrate (Fe(NO3)3•9H2O) to form [Fe(SCN)(H2O)5]2+, which absorbs at 460 nm. (b–d) Mechanisms of _S_-sulfhydryl formation. Protein _S_-sulfhydryls can form upon the reaction of H2S with (b) a protein sulfenic acid or (c) a disulfide, though the latter is very slow owing to the poor reductant activity of H2S. (d) Alternatively, _S_-sulfhydryls can form from nucleophilic attack of a protein thiolate on H2S2. (e) Once formed, protein _S_-sulfhydryls can be shuttled between proteins via transsulfhydration, as is well established for the sulfurtransferase IscS (eq 1). Reaction of a protein _S_-sulfhydryl with a second cysteine can alternatively yield a disulfide (eq 2). Akin to _S_-nitrosothiols, the relative p_K_a of the protein and H2S thiols will influence which reaction occurs.
Chart 17. Protein _S_-Sulfhydryls React with NEM (Equation 1), IAM (Equation 2), and MMTS (Equation 3) to Yield the Corresponding Di- or Trisulfides
Figure 20
Indirect chemical methods to detect _S_-sulfhydryls. (a) Free thiols (blue) and _S_-sulfhydryls (purple) are modified with a red fluorescent maleimide reagent. The disulfide linked maleimide adducts derived from _S_-sulfhydryls are reduced by DTT. Protein modification by S-sulfhydration is detected as a decrease in signal by in-gel fluorescence. (b) Indirect method to simultaneously monitor _S_-sulfhydryl and _S_-nitrosothiol formation. Free thiols (blue) and _S_-sulfhydryls (purple) are modified with a red fluorescent maleimide reagent. _S_-nitrosothiols (green) are reduced with ascorbate and nascent thiols modified with a green fluorescent maleimide reagent. The disulfide linked maleimide adducts derived from _S_-sulfhydryls are reduced by DTT. Protein modification by S-sulfhydration and S-nitrosylation are detected as a decrease in red signal and increase in green signal by in-gel fluorescence, respectively. (c) Free thiols (blue) and _S_-sulfhydryls (purple) are modified by IAM. The disulfide-linked alkylation adduct and all other reversible cysteine modifications (green) are reduced with DTT and nascent thiols are modified with IAP-biotin. Cysteine oxidation is monitored as an increase in signal by avidin blot.
Similar articles
- Nanotransducers in cellular redox signaling: modification of thiols by reactive oxygen and nitrogen species.
Cooper CE, Patel RP, Brookes PS, Darley-Usmar VM. Cooper CE, et al. Trends Biochem Sci. 2002 Oct;27(10):489-92. doi: 10.1016/s0968-0004(02)02191-6. Trends Biochem Sci. 2002. PMID: 12368076 Review. - Cysteine-mediated redox signalling in the mitochondria.
Bak DW, Weerapana E. Bak DW, et al. Mol Biosyst. 2015 Mar;11(3):678-97. doi: 10.1039/c4mb00571f. Epub 2014 Dec 18. Mol Biosyst. 2015. PMID: 25519845 Review. - A direct way of redox sensing.
Benoit R, Auer M. Benoit R, et al. RNA Biol. 2011 Jan-Feb;8(1):18-23. doi: 10.4161/rna.8.1.13555. Epub 2011 Jan 1. RNA Biol. 2011. PMID: 21220941 - Reversible cysteine oxidation in hydrogen peroxide sensing and signal transduction.
García-Santamarina S, Boronat S, Hidalgo E. García-Santamarina S, et al. Biochemistry. 2014 Apr 29;53(16):2560-80. doi: 10.1021/bi401700f. Epub 2014 Apr 16. Biochemistry. 2014. PMID: 24738931 Review. - [Chemical approaches for trapping protein thiols and their oxidative modification].
Huang CS, Zhu WP, Xu YF, Qian XH. Huang CS, et al. Yao Xue Xue Bao. 2012 Mar;47(3):280-90. Yao Xue Xue Bao. 2012. PMID: 22645750 Review. Chinese.
Cited by
- Lifestyle-specific S-nitrosylation of protein cysteine thiols regulates Escherichia coli biofilm formation and resistance to oxidative stress.
Barraud N, Létoffé S, Beloin C, Vinh J, Chiappetta G, Ghigo JM. Barraud N, et al. NPJ Biofilms Microbiomes. 2021 Apr 13;7(1):34. doi: 10.1038/s41522-021-00203-w. NPJ Biofilms Microbiomes. 2021. PMID: 33850153 Free PMC article. - The Human 2-Cys Peroxiredoxins form Widespread, Cysteine-Dependent- and Isoform-Specific Protein-Protein Interactions.
van Dam L, Pagès-Gallego M, Polderman PE, van Es RM, Burgering BMT, Vos HR, Dansen TB. van Dam L, et al. Antioxidants (Basel). 2021 Apr 20;10(4):627. doi: 10.3390/antiox10040627. Antioxidants (Basel). 2021. PMID: 33923941 Free PMC article. - Ammonia Toxicity and Associated Protein Oxidation: A Single-Cell Surface Enhanced Raman Spectroscopy Study.
Redolfi-Bristol D, Mangiameli A, Yamamoto K, Marin E, Zhu W, Mazda O, Riello P, Pezzotti G. Redolfi-Bristol D, et al. Chem Res Toxicol. 2024 Jan 15;37(1):117-125. doi: 10.1021/acs.chemrestox.3c00368. Epub 2023 Dec 26. Chem Res Toxicol. 2024. PMID: 38146714 Free PMC article. - ROS signaling and redox biology in endothelial cells.
Panieri E, Santoro MM. Panieri E, et al. Cell Mol Life Sci. 2015 Sep;72(17):3281-303. doi: 10.1007/s00018-015-1928-9. Epub 2015 May 14. Cell Mol Life Sci. 2015. PMID: 25972278 Free PMC article. Review. - An Excited State Intramolecular Proton Transfer-Based Fluorescent Probe with a Large Stokes Shift for the Turn-on Detection of Cysteine: A Detailed Theoretical Exploration.
Ding S, Xu A, Sun A, Xia Y, Liu Y. Ding S, et al. ACS Omega. 2020 Jul 31;5(31):19695-19701. doi: 10.1021/acsomega.0c02393. eCollection 2020 Aug 11. ACS Omega. 2020. PMID: 32803064 Free PMC article.
References
- Hirooka Y.; Sagara Y.; Kishi T.; Sunagawa K. Circ. J. 2010, 74, 827. - PubMed
- Koh C. H.; Whiteman M.; Li Q. X.; Halliwell B.; Jenner A. M.; Wong B. S.; Laughton K. M.; Wenk M.; Masters C. L.; Beart P. M.; Bernard O.; Cheung N. S. J. Neurochem. 2006, 98, 1278. - PubMed
- Henriksen E. J.; Diamond-Stanic M. K.; Marchionne E. M. Free Radical Biol. Med. 2011, 51, 993. - PMC - PubMed
- Suh Y. A.; Arnold R. S.; Lassegue B.; Shi J.; Xu X.; Sorescu D.; Chung A. B.; Griendling K. K.; Lambeth J. D. Nature 1999, 401, 79. - PubMed
- Grek C. L.; Tew K. D. Curr. Opin. Pharmacol. 2010, 10, 362. - PMC - PubMed
- Jacob C.; Winyard P. G.. Introduction in Redox Signaling and Regulation in Biology and Medicine; Jacob C., Winyard P. G., Eds.; Wiley-VCH: Weinheim, Germany, 2009; pp 1–12.
- Reddie K. G.; Carroll K. S. Curr. Opin. Chem. Biol. 2008, 12, 746. - PubMed
- Finkel T. Sci. Signalinging 2012, 5, pe10. - PubMed
- Fratelli M.; Demol H.; Puype M.; Casagrande S.; Eberini I.; Salmona M.; Bonetto V.; Mengozzi M.; Duffieux F.; Miclet E.; Bachi A.; Vandekerckhove J.; Gianazza E.; Ghezzi P. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 3505. - PMC - PubMed
- Hess D. T.; Matsumoto A.; Kim S. O.; Marshall H. E.; Stamler J. S. Nat. Rev. Mol. Cell Biol. 2005, 6, 150. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources